Research
General overview and Research Groups
The Turing Centre aims to stimulate a highly interdisciplinary community, especially physicists, mathematicians and computer scientists who, together with biologists, provide the means to base experimental studies on solid quantitative grounds, to identify new questions and conceptualize new ways of understanding the complexity of living systems.
Life is based on dynamic systems whose organisational principles remain unclear due to their complexity. Such complexity has different origins. Living systems are the product of a long evolutionary history and are intrinsically dynamic as they develop, renew their components, adapt to their physiological environments and are trained. This is illustrated for instance in the development of embryos and organs, and in the function of the immune and nervous systems.
Understanding biological complexity is one of the most significant challenges for the future. It will impact both basic research and medicine, since diseases emerge from anomalies in the organisation and dynamics of living systems.
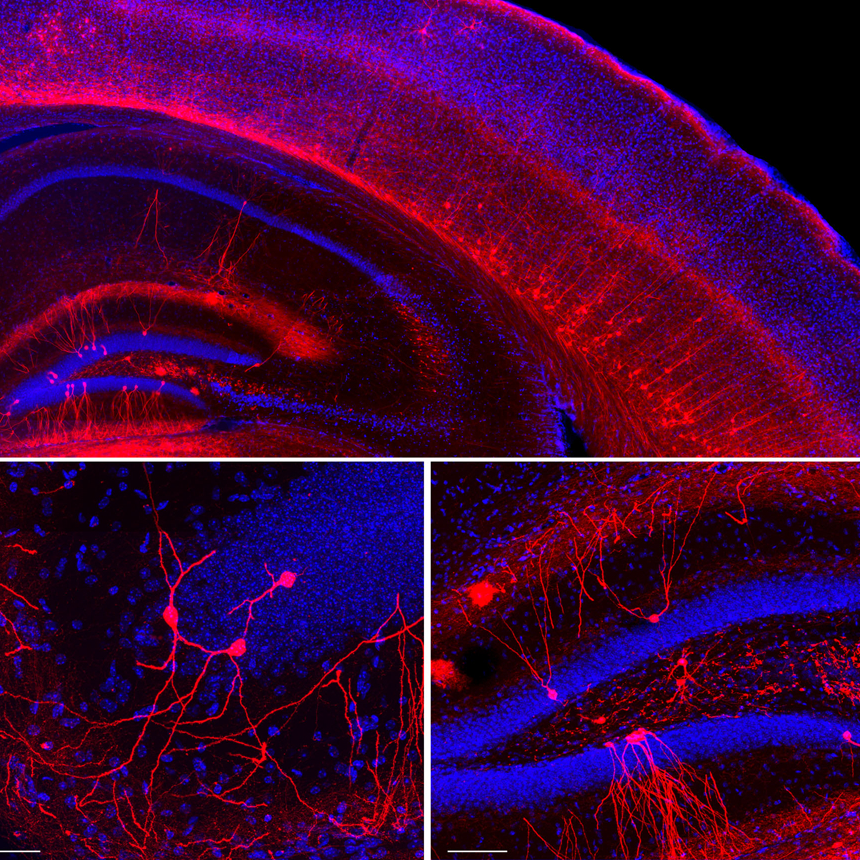
The Turing Centre addresses the nature of the information underlying biological processes, and the organisation and dynamics of biological structures. Five over-arching questions bring together biologists interested in development, neuroscience and immunology experts along with physicists, while computational biologists and mathematicians underlie the research effort:
How does genetic information impact cell fates?
How do signalling dynamics impact cell differentiation?
How is information encoded in collective cellular dynamics?
How do networks of cell interactions form and function collectively?
How does mechanics impact cell and tissue organisation and dynamics?
CENTURI recruits researchers and post-docs with backgrounds in computational biology, theoretical physics or biophysics, with a strong interest in working in a cross-disciplinary environment.
CENTURI fellows
Principal Investigators
Mathematical modelling of the ecology & evolution of host-associated microbiota
Brain mechanisms of flexible behavior
Understanding developmental reproducibility at the single cell scale
Self-organization and collective effects in living systems
Neural networks for the representation of 3D environments
Modeling and Measuring Soft Living Matter
Phillipe Roudot (Institut Fresnel)
Dynamic bioimage mining for live cell microscopy
Out-of-equilibrium Mechanics (OM)
Learning Meaningful Representations of Life
Florence Bansept (LCB) - Mathematical modelling of the ecology & evolution of host-associated microbiota
Lorenzo Fontolan (Inmed - CPT) - Brain mechanisms of flexible behavior
Léo Guignard (LIS) - Understanding developmental reproducibility at the single cell scale
Matthias Merkel (CPT) - Self-organization and collective effects in living systems
Hervé Rouault (CPT - INMED) - Neural networks for the representation of 3D environments
Pierre Ronceray (CPT) -Theoretical modeling and inference methods for the physics of soft biological matter
Phillipe Roudot (Institut Fresnel) - Dynamic bioimage mining for live cell microscopy
Jean-François Rupprecht (CPT) - Out-of-equilibrium Mechanics (OM)
Paul Villoutreix (LIS) - Marseille Machine Learning Team
Postdoctoral fellow
Mehdi Abbasi (CINaM, M2P2) -Understanding margination, from vessels to vascular networks
Ludovica Bachschmid Romano (Inmed) - Computational frameworks to link structure, dynamics and function in large-scale neural circuits
Maximilien Courgeon (IBDM, CPT) - Self-organising principles of Class IV neuron dendritic arborization in Drosophila
Paulina Garcia Gonzalez (IBDM) - EPIC-Mitochondria
Gesa Loof (LIS) - Early drivers of symmetry breaking in pre-implantation mouse embryo cell fate specification
Florian Labourel (LCB) - Metabolism and multi-stable equilibria in the gut microbiome
Sujit Kumar Nath (IBDM, CPT) - The effect of changing curvature on tissue flows in Drosophila embryo
Vitor Marquioni Monteiro (LCB) - Mathematical Modeling of Gut Microbiome
Ying Zhou (Inserm, TAGC) - Quantitive micromethods for rapid characterization of immune cells activity in infection and cancerous context
Understanding margination, from vessels to vascular networks
Ludovica Bachschmid Romano (Inmed)
Computational frameworks to link structure, dynamics and function in large-scale neural circuits
Maximilien Courgeon (IBDM/CPT)
Self-organising principles of Class IV neuron dendritic arborization in Drosophila
Paulina Garcia Gonzalez (CIML/IBDM)
EPIC-Mitochondria
Metabolism and multi-stable equilibria in the gut microbiome
Early drivers of symmetry breaking in pre-implantation mouse embryo cell fate specification
Quantitive micromethods for rapid characterization of immune cells activity in infection and cancerous context
PhD students
Lorna Ammer (LAI, CIML) - The impact of dynamic interactions between meningeal immune cells on neurodevelopment
Ahmad Awada (LAI, I2M) - Reverse adhesive haptotaxis: from reductionist experimentation to mathematical and physical modeling
Lama Awada (CIML, LAI) - Imprint of mechanical forces on antibody affinity maturation in B cell immune responses
Alexandre Bonomo - Multi-view machine learning analysis of intestinal immune response
Nadine Ben Boina (I2M, MMG) - Mathematical Modelling of rare diseases
Marie-Jose Chaaya (I2M, IBDM) Modeling tumor innervation: impact on immune cells and anti-cancer treatment
Jessica Chevallier (CIML, TAGC, LAI) - Spatial mapping of gene expression to decipher the thymic cellular
Arthur Coët - Deciphering bacterial colonization of marine snow
Marie Dessard - How cholesterol and phospholipids fine tune T cell early mechanosensitivity?
Jaime Fernandez MacGregor (TAGC) - Modeling human network perturbations by bacterial proteins
Andonis Gerardos (CINaM) - How to learn a stochastic partial differential equation from a video?
Alice Gros (IBDM, LIS) - Morphogenesis variability in embryonic organoids.
Mathias Ramm Haugland (IBDM/LIS) - MitoLearner: deep learning and explainable AI to understand the role of mitochondria genes in cancer cells
Andrei Karpov - Physical and structural characterization of the interplay between the cell membranes, actin filaments and Chikungunya protein nsP1
Martin Lardy (IRPHE,IBDM) - Computational Fluid Dynamics to infer embryonic tissue rheology
Emma Legait - Deciphering the self-organizing principles governing the cellular composition and growth of tumors
Marvin Leria (IBDM) - PCP plasticity in the basal metazoan Trichoplax adhaerens
Ismahene Mesbah (LAI, IBDM) - Decoding protein structural folds, sequence and structural motifs
Piyush Mishra (I2M, Institut Fresnel) - Weakly-supervised inferences of molecular dynamics for fluorescence imaging in physiological environments
Alexandre Ortega - Development of acoustic techniques to study the biophysical properties of cell clusters
Harshit Pateria (IBDM/MMG) - Integrating transcription and lineage to define spatiotemporal trajectories of cardiac progenitor cells
Jessica Pereira Silva (INMED/I2M) - Behavioural grammar and synaptic rules of the social brain
Marc-Eric Perrin (IBDM, CPT) - Models of neuronal growth during embryogenesis as self-interacting dynamical trees
Devam Purohit (CINaM, I2M) - The muscle spring: quantifying and manipulating the molecular elasticity of muscle in vitro and in vivo
Florian Saby (IRPHE/LCB) - Understanding Decision-Making in Myxococcus xanthus
Mirindra Ratsifandrihamanana (INMED, CPT) - Circuits supporting dynamics transitions in the developing hippocampus
Hilde Langengen Teigen (INMED, INT) Neuromodulatory mechanisms of predictive processing in the mouse visual cortex
Rémy Torro (CINaM, LAI) - Mapping and classifying the topography of adhesive cell interface with high precision
Paul Uteza (CPT, INMED) - Neural networks for the representation of 3D environments
João Pedro Valeriano Miranda (CINaM) - Learning hidden structures in the stochastic dynamics of living systems
Jules Vanaret (I2M, IBDM) - OrganoDyn - Organoid dynamic reconstruction across scales
The impact of dynamic interactions between meningeal immune cells on neurodevelopment
Reverse adhesive haptotaxis: from reductionist experimentation to mathematical and physical modeling
Imprint of mechanical forces on antibody affinity maturation in B cell immune responses
Multi-view machine learning analysis of intestinal immune response
Mathematical Modelling of rare diseases
Modeling tumor innervation: impact on immune cells and anti-cancer treatment
Jessica Chevallier (CIML/TAGC/LAI)
Spatial mapping of gene expression to decipher the thymic cellular
Deciphering bacterial colonization of marine snow
How cholesterol and phospholipids fine tune T cell early mechanosensitivity?
Jaime Fernandez MacGregor (TAGC/AFMB)
Modeling human network perturbations by bacterial proteins
How to learn a stochastic partial differential equation from a video?
Morphogenesis variability in embryonic organoids.
Mathias Ramm Haugland (IBDM/LIS)
MitoLearner: deep learning and explainable AI to understand the role of mitochondria genes in cancer cells
Physical and structural characterization of the interplay between the cell membranes, actin filaments and Chikungunya protein nsP1
Deciphering the self-organizing principles governing the cellular composition and growth of tumors
PCP plasticity in the basal metazoan Trichoplax adhaerens
Decoding protein structural folds, sequence and structural motifs
Piyush Mishra (I2M/Institut Fresnel)
Weakly-supervised inferences of molecular dynamics for fluorescence imaging in physiological environments
Alexandre Ortega (LMA/LAI/IUSTI)
Development of acoustic techniques to study the biophysical properties of cell clusters
Integrating transcription and lineage to define spatiotemporal trajectories of cardiac progenitor cells
Jessica Pereira Silva (INMED/I2M)
Behavioural grammar and synaptic rules of the social brain
Models of neuronal growth during embryogenesis as self-interacting dynamical trees
The muscle spring: quantifying and manipulating the molecular elasticity of muscle in vitro and in vivo
Mirindra Ratsifandri-hamanana (INMED/INT)
Circuits supporting dynamics transitions in the developing hippocampus
Understanding Decision-Making in Myxococcus xanthus
Hilde Langengen Teigen (INMED/ INT)
Neuromodulatory mechanisms of predictive processing in the mouse visual cortex
Mapping and classifying the topography of adhesive cell interface with high precision
Neural networks for the representation of 3D environments
João Pedro Valeriano Miranda (CINaM)
Learning hidden structures in the stochastic dynamics of living systems
Engineers
Lea Chabot - Bioinformatics Engineer
Audrey Comte - Software Development Engineer
Fabrice Dessolis - Mechatronics Engineer
Thang Duong Quoc Le - Image Data Processing & Analysis
Mai Han Hoang - Image Data Processing & Analysis
Mathias Lechelon - Mechatronics
Stefania Sarno - Data Neuroscientist
Thomas Vannier - Bioinformatics Data Analyst
Mechatronics
Alumni
Since 2017, talented and innovative scientists have helped us shape CENTURI and collaborate on federating a diverse community and to ensure its commitment to an interdiciplinary approach of Science and Research.
Photography by ©Criscuolo
Publications
2024
- Membrane tilt drives phase separation of adhesion receptors. S.-Z. Lin, R. Changede, A. J. Farrugia, A. D. Bershadsky, M. P. Sheetz, J. Prost, J.-F. Rupprecht, Physical Review Letters, https://journals.aps.org/prl/abstract/10.1103/PhysRevLett.132.188402
- PatternJ: an ImageJ toolset for the automated and quantitative analysis of regular spatial patterns found in sarcomeres, axons, somites, and more.
- Dissolved organic matter offsets the detrimental effects of climate change in the nitrogen-fixing cyanobacterium Crocosphaera. Alba Filella, Jacqueline Umbricht, Angelina Klett, Angela Vogts, Thomas Vannier, Olivier Grosso, Maren Voss, Lasse Riemann, Mar Benavides. Limnology and Oceanography Letters 2024 Mar 5. doi: https://doi.org/10.1002/lol2.10380.
2023
- u-track3D: Measuring, navigating, and validating dense particle trajectories in three dimensions. Roudot et al.,. Cell Reports Methods, vol. 3, no. 12, p. 100655, Dec. 2023, doi: 10.1016/j.crmeth.2023.100655.
- A Detector-Independent Quality Score for Cell Segmentation Without Ground Truth in 3D Live Fluorescence Microscopy. Vanaret, V. Dupuis, P.-F. Lenne, F. Richard, S. Tlili, and P. Roudot. IEEE Journal of Selected Topics in Quantum Electronics, vol. 29, no. 4: Biophotonics, pp. 1–12, Jul. 2023, doi: 10.1109/JSTQE.2023.3275108.
- Curvature-induced cell rearrangements in biological tissues. Y. Lou, S. Theis, J.-F. Rupprecht, T. Hiraiwa, T. E. Saunders
Physical Review Letters (2023) DOI:10.1103/PhysRevLett.130.108401 https://journals.aps.org/prl/abstract/10.1103/PhysRevLett.130.108401 - Structure and Rheology in Vertex Models under Cell-Shape-Dependent Active Stresses. S.-Z. Lin, M. Merkel, J.-F. Rupprecht Physical Review Letters (2023) DOI:10.1103/PhysRevLett.130.058202 https://journals.aps.org/prl/abstract/10.1103/PhysRevLett.130.058202
- pH gradients guide ADF/cofilin isoforms in pollen tubes. Dinet C., Michelot A. Journal of Cell Biology (JCB) 222(11), 2023 Oct, DOI:10.1083/jcb.202310012
- Reconstitution of actin-based cellular processes: Why encapsulation changes the rules. Kandiyoth FB, Michelot A. Eur J Cell Biol. 2023 Dec;102(4):151368. doi: 10.1016/j.ejcb.2023.151368. Epub 2023 Oct 30. PMID: 37922812.
- Protocol to image and analyze hippocampal network dynamics in non-anesthetized mouse pups. Ratsifandrihamanana, M. R., Dard, R. F., Denis, J., Cossart, R. & Picardo, M. A. STAR Protoc. 4, 102760 (2023). DOI: 10.1016/j.xpro.2023.102760
- Extrinsic control of the early postnatal CA1 hippocampal circuits. Leprince, E. et al. Neuron 111, 888-902.e8 (2023). d
- Cortical neuronal assemblies coordinate with EEG microstate dynamics during resting wakefulness. Boyce, R., Dard, R. F. & Cossart, R. Cell Rep. 42, 112053 (2023). DOI: 10.1016/j.celrep.2023.112053
- Classification of red cell dynamics with convolutional and recurrent neural networks: a sickle cell disease case study. M. Darrin, A. Samudre, M. Sahun, S. Atwell, C. Badens, A. Charrier, E. Helfer, A. Viallat, V. Cohen-Addad, and S. Giffard-Roisin, Scientific Reports 13, 745 (2023). https://doi.org/10.1038/s41598-023-27718-w
- Light-inducible T cell engagers trigger, tune, and shape the activation of primary T cells. Morgane Jaeger, Amandine Anastasio, Léa Chamy, Sophie Brustlein, Renaud Vincentelli, Fabien Durbesson, Julien Gigan, Morgane Thépaut, Rémy Char, Maud Boussand, Mathias Lechelon, Rafael J. Argüello, Didier Marguet, Hai-Tao He, Rémi Lasserre. PNAS 2023 Sep 18. doi: https://doi.org/10.1101/2022.04.15.488452.
- Prolonged dysbiosis and altered immunity under nutritional intervention in a physiological mouse model of severe acute malnutrition. Fanny Hidalgo-Villeda, Matthieu Million, Catherine Defoort, Thomas Vannier, Ljubica Svilar, Margaux Lagier, Camille Wagner, Cynthia Arroyo-Portilla, Lionel Chasson, Cécilia Luciani, Vincent Bossi, Jean-Pierre Gorvel, Hugues Lelouard, Julie Tomas. iScience 2023 jun 16. doi: https://doi.org/10.1016/j.isci.2023.106910
- Epithelial disruption drives mesendoderm differentiation in human pluripotent stem cells by enabling TGF-β protein sensing. Thomas Legier, Diane Rattier, Jack Llewellyn, Thomas Vannier, Benoit Sorre, Flavio Maina & Rosanna Dono. Nature Communications 2023 Jan 21. doi: https://doi.org/10.1038/s41467-023-35965-8.
- Meisosomes, folded membrane microdomains between the apical extracellular matrix and epidermis. Dina Aggad, Nicolas Brouilly, Shizue Omi, Clara Luise Essmann, Benoit Dehapiot, Cathy Savage-Dunn, Fabrice Richard, Chantal Cazevieille, Kristin A Politi, David H Hall, Remy Pujol, Nathalie Pujol. eLife 2023 Mar 13. doi: https://doi.org/10.7554/eLife.75906
- Probing mechanical interaction of immune receptors and cytoskeleton by membrane nanotube extraction. Manca, F., Eich, G., N’Dao, O. et al. Sci Rep 13, 15652 (2023). https://doi.org/10.1038/s41598-023-42599-9
- Pathogen metabolite checkpoint: NHR on guard. Immunity 2023 Apr 11;56(4):744-746.
- Curvature gradient drives polarized tissue flow in the Drosophila embryo. February 1, 2023 120 (6) e2214205120 https://doi.org/10.1073/pnas.2214205120
- Structure and Rheology in Vertex Models under Cell-Shape-Dependent Active Stresses. Shao-Zhen Lin, Matthias Merkel, and Jean-François Rupprecht. Phys. Rev. Lett. 130, 058202, https://doi.org/10.1103/PhysRevLett.130.058202
- Soft matter roadmap. Jean-Louis Barrat, Emanuela Del Gado, Stefan U Egelhaaf, Matthias Merkel, Pierre Ronceray and Jean-François Rupprecht. , , DOI 10.1088/2515-7639/ad06cc
- Influence of storage and buffer composition on the mechanical behavior of flowing red blood cells. Merlo A, Losserand S, Yaya F, Connes P, Faivre M, Lorthois S, Minetti C, Nader E, Podgorski T, Renoux C, Coupier G, Franceschini E, Biophys J. 2023; 122(2):360-373. doi:10.1016/j.bpj.2022.12.005
- Hyper-cores promote localization and efficient seeding in higher-order processes. Marco Mancastroppa, Iacopo Iacopini, Giovanni Petri, Alain Barrat. Commun. 14, 6223 (2023) https://doi.org/10.1038/s41467-023-41887-2
- Distinguishing simple and complex contagion processes on networks. Giulia Cencetti, Diego Andrés Contreras, Marco Mancastroppa, Alain Barrat. Phy. Rev. Lett. 130, 247401 (2023). https://doi.org/10.1103/PhysRevLett.130.247401
- Contact networks have small metric backbones that maintain community structure and are primary transmission subgraphs. Rion Brattig Correia, Alain Barrat, Luis M. Rocha. PLoS Comput Biol 19(2): e1010854 (2023) https://doi.org/10.1371/journal.pcbi.1010854
- Inferring cell cycle phases from a partially temporal network of protein interactions. Maxime Lucas, Arthur Morris, Alex Townsend-Teague, Laurent Tichit, Bianca H Habermann, Alain Barrat. Cell Reports Methods 3, 100397 (2023) https://doi.org/10.1016/j.crmeth.2023.100397
- A gut meta-interactome map reveals modulation of human immunity by microbiome effectors. ,
- mimicINT: a workflow for microbe-host protein interaction inference.
- Modulation of human kinase activity through direct interaction with SARS-CoV-2 proteins.
- Spatiotemporal transcriptomic maps of whole mouse embryos at the onset of organogenesis. Sampath Kumar, A., Tian, L., Bolondi, A., Guignard L. et al. Nat Genet 55, 1176–1185 (2023). https://doi.org/10.1038/s41588-023-01435-6
- Random Walk with Restart on multilayer networks: from node prioritisation to supervised link prediction and beyond.
- Zoo Guide to Network Embedding. A. Baptista, R. J. Sanchez-Garcia, A. Baudot, G. Bianconi. Arxiv. https://doi.org/10.48550/arXiv.2305.03474
- Transit time theory for a droplet passing through a slit in pressure-driven low Reynolds-number flows. S. W. Borbas, K. Shen, C. Ji, A. Viallat, E. Helfer, Z. Peng. Micromachines 14, 2040 (2023). https://doi.org/10.3390/mi14112040
- Physical mechanisms of red blood cell splenic filtration. A. Moreau, F. Yaya, H. Lu, A. Surendranath, A. Charrier, B. Dehapiot, E. Helfer, A. Viallat, Z. Peng. Proceedings of the National Academy of Sciences 120, e2300095120 (2023). https://doi.org/10.1073/pnas.2300095120
- ShIVA: a user-friendly and interactive interface giving biologists control over their single-cell RNA-seq data. Rudy Aussel, Muhammad Asif, Sabrina Chenag, Sébastien Jaeger, Pierre Milpied & Lionel Spinelli. Sci Rep 13, 14377 (2023). https://doi.org/10.1038/s41598-023-40959-z
- Enhanced cell viscosity: A new phenotype associated with lamin A/C alterations. Cécile Jebane, Alice-Anaïs Varlet, Marc Karnat, Lucero M. Hernandez- Cedillo, Amélie Lecchi, Frédéric Bedu, Camille Desgrouas, Corinne Vigouroux, Marie-Christine Vantyghem, Annie Viallat, Jean-François Rupprecht, Emmanuèle Helfer, Catherine Badens, iScience Volume 26, ISSUE 10, 107714. https://doi.org/10.1016/j.isci.2023.107714
- Body stiffness is a mechanical property that facilitates contact-mediated mate recognition in Caenorhabditis elegans. Jen-Wei Weng, Heenam Park, Claire Valotteau, Rui-Tsung Chen, Clara L. Essmann, Nathalie Pujol, Paul W. Sternberg, Chun-Hao Chen. Curr Biol. 2023 Jul 28;S0960-9822(23)00927-2. https://doi.org/10.1016/j.cub.2023.07.020
- Prominent in vivo influence of single interneurons in the developing barrel cortex. Bollmann, Y., Modol, L., Tressard, T., Cossart, R., et al., Nat Neurosci 26, 1555–1565 (2023). https://doi.org/10.1038/s41593-023-01405-5
- Alcanivorax borkumensis biofilms enhance oil degradation by interfacial tubulation. M. Prasad, N. Obana, S.-Z. Lin, Jean-François Rupprecht et al., Science 381, 748-753 (2023). DOI: 10.1126/science.adf3345
- Analytical theory for a droplet squeezing through a circular pore in creeping flows under constant pressures. Zhengxin Tang (唐正昕), François Yaya, Ethan Sun, Lubna Shah, Jie Xu, Annie Viallat, Emmanuèle Helfer, Zhangli Peng (彭张立). Physics of Fluids 35, 082016 (2023), https://doi.org/10.1063/5.0156349
- Talin and kindlin cooperate to control the density of integrin clusters. Pernier J, Cardoso Dos Santos M, Souissi M, Joly A, Narassimprakash H, Rossier O, Giannone G, Helfer E, Sengupta K, Le Clainche C. J Cell Sci. 2023 Apr 15;136(8):jcs260746. doi: 10.1242/jcs.260746. Epub 2023 Apr 21. PMID: 37083041.
- Inferring cell cycle phases from a partially temporal network of protein interactions. Maxime Lucas, Arthur Morris, Alex Townsend-Teague, Laurent Tichit, Bianca Habermann, Alain Barrat. Cell Reports Methods, Volume 3, Issue 2, 2023, 100397, ISSN 2667-2375, https://doi.org/10.1016/j.crmeth.2023.100397.
- Growth anisotropy of the extracellular matrix shapes a developing organ. Harmansa, S., Erlich, A., Eloy, C. et al. Growth anisotropy of the extracellular matrix shapes a developing organ. Nat Commun 14, 1220 (2023). https://doi.org/10.1038/s41467-023-36739-y
- Two-point optical manipulation reveals mechanosensitive remodeling of cell–cell contacts in vivo. Kenji Nishizawa, Shao-Zhen Lin, Claire Chardès, Jean-François Rupprecht and Pierre-François Lenne. PNAS, March 22, 2023, 120 (13) e2212389120, https://doi.org/10.1073/pnas.221238912
- Deforming polar active matter in a scalar field gradient. Muhamet Ibrahimi1 and Matthias Merkel1, New Journal of Physics, 13 January 2023, DOI 10.1088/1367-2630/acb2e5
2022
- Labelling of Active Transcription Sites with Argonaute NRDE-3-Image Active Transcription Sites in vivo in Caenorhabditis elegans. Bio Protoc. 2022 June5 12(11):e4427. doi: 10.21769/BioProtoc.4427.
- PRC1 chromatin factors strengthen the consistency of neuronal cell fate specification and maintenance in C. elegans. Guillaume Bordet, Carole Couillault, Fabien Soulavie, Konstantina Filippopoulou, Vincent Bertrand. Plos Genetics, 2022. https://doi.org/10.1371/journal.pgen.1010209
- Mechanical stress driven by rigidity sensing governs epithelial stability. S. Sonam(1), L. Balasubramaniam(1), S.-Z. Lin(1), ..., J.-F. Rupprecht, B. Ladoux Nature Physics (2022)
- Active nematic flows on curved surfaces. S. Bell (1), S.-Z. Lin(1), J.-F. Rupprecht, J. Prost ,Physical Review Letters (2022)
- Non-linear elastic properties of actin patches to partially rescue yeast endocytosis efficiency in the absence of the cross-linker Sac6. Soft Matter, 2022,18, 1479-1488, DOI https://doi.org/10.1039/D1SM01437D (correction in doi: 10.1039/d2sm90083a)
- Specialization of actin isoforms derived from the loss of key interactions with regulatory factors. Boiero Sanders M, Toret CP, Guillotin A, Antkowiak A, Vannier T, Robinson RC, Michelot A. EMBO J. 2022 Mar 1;41(5):e107982. doi: 10.15252/embj.2021107982
- The cellular slime mold Fonticula alba forms a dynamic, multicellular collective while feeding on bacteria. Christopher Toret, Andrea Picco, Micaela Boiero-Sanders, Alphee Michelot, Marko Kaksonen, Current Biology, Volume 32, Issue 9, 2022, Pages 1961-1973.e4, https://doi.org/10.1016/j.cub.2022.03.018.
- Enhanced wild-type MET receptor levels in mouse hepatocytes attenuates insulin-mediated signalling. Rada P.*, Lamballe F., Carceller-López E., Hitos A.B., Sequera C., Maina F.*, Valverde A.M.* Cells, Feb 24;11(5):793 (2022). PMID: 35269415. *shared corresponding authorship.MYC and MET cooperatively drive hepatocellular carcinoma with distinct molecular traits and vulnerabilities. Sequera C., Grattarola M., Holczbauer A., Dono R., Pizzimenti S., Barrera G., Wangensteen K., Maina F. Cell Death and Disease, 24;13(11):994 (2022). PMID: 36433941.
- Time-limited alterations in cortical activity of a knock-in mice model of KCNQ2-related developmental and epileptic encephalopathy. Najoua Biba, Hélène Becq, Emilie Pallesi-Pocachard, Stefania Sarno, Samuel Granjeaud, Aurélie Montheil, Marie Kurz, Laurent Villard, Mathieu Milh, Pierre-Pascal Lenck Santini, Laurent Aniksztejn. The Journal of Physiology 2022 Apr 7. doi: https://doi.org/10.1113/JP282536
- Un nouveau modèle murin de la malnutrition aiguë sévère infantile révèle un défaut de l’immunité de la plaque de Peyer ainsi qu’une altération du microbiote et du profil métabolomique. F. Hidalgo-Villeda, M. Million, C. Defoort, F. Sicard, M. Lagier, C. Wagner, C. Arroyo-Portilla, L. Chasson, Thomas Vannier, J.-P. Gorvel, H. Lelouard, J. Tomas. Nutrition Clinique et Métabolique 2022 Feb. doi: DOI: 10.1016/j.nupar.2021.12.012
- Specialization of actin isoforms derived from the loss of key interactions with regulatory factors. Micaela Boiero Sanders, Christopher P. Toret, Audrey Guillotin, Adrien Antkowiak, Thomas Vannier, Robert C. Robinson and Alphée Michelot. EMBO J 2022 Feb 18;:e107982. doi: 10.15252/embj.2021107982
- Stiffening of under-constrained spring networks under isotropic strain.
- Phase separation dynamics in deformable droplets.
- Implementation of cellular bulk stresses in vertex models of biological tissues. Lin, SZ., Merkel, M. & Rupprecht, JF. Eur. Phys. J. E 45, 4 (2022). https://doi.org/10.1140/epje/s10189-021-00154-2
- Longitudinal to Transverse Metachronal Wave Transitions in an In Vitro Model of Ciliated Bronchial Epithelium. Olivier Mesdjian, Chenglei Wang, Simon Gsell, Umberto D’Ortona, Julien Favier, Annie Viallat, and Etienne Loiseau. Phys. Rev. Lett. 129, 038101. https://doi.org/10.1103/PhysRevLett.129.038101
- Minimizing school disruption under high incidence conditions due to the Omicron variant in France, Switzerland, Italy, in January 2022. Elisabetta Colosi, Giulia Bassignana, Alain Barrat, Bruno Lina, Philippe Vanhems, Julia Bielicki, Vittoria Colizza. Euro Surveill. 2023;28(5):pii=2200192. https://doi.org/10.2807/1560-7917.ES.2023.28.5.2200192
- Impact of contact data resolution on the evaluation of interventions in mathematical models of infectious diseases. Diego A Contreras, Elisabetta Colosi, Giulia Bassignana, Vittoria Colizza, Alain Barrat. J. R. Soc. Interface 19:20220164 (2022) https://doi.org/10.1098/rsif.2022.0164
- The temporal rich club phenomenon. Nicola Pedreschi, Demian Battaglia, Alain Barrat. Nat. Phys. 18:931 (2022) https://doi.org/10.1038/s41567-022-01634-8
- Screening and vaccination against COVID-19 to minimise school closure: a modelling study. Elisabetta Colosi, Giulia Bassignana, Diego A Contreras, Canelle Poirier, Pierre-Yves Boëlle, Simon Cauchemez, Yazdan Yazdanpanah, Bruno Lina, Arnaud Fontanet, Alain Barrat, Vittoria Colizza. Lancet Infect Dis 22:977 (2022) https://doi.org/10.1016/S1473-3099(22)00138-4
- Transition from simple to complex contagion in collective decision-making. Nikolaj Horsevad, David Mateo, Robert E. Kooij, Alain Barrat, Roland Bouffanais. Nature Communications 13:1442 (2022) https://doi.org/10.1038/s41467-022-28958-6
- In Depth Exploration of the Alternative Proteome of Drosophila melanogaster. Bertrand Fabre, Sebastien A. Choteau, Carine Duboé, Christine Brun et al. Front. Cell Dev. Biol., Volume 10 - 2022. https://doi.org/10.3389/fcell.2022.901351
- Epi-MEIF: detecting higher order epistatic interactions for complex traits using mixed effect conditional inference forests. Saswati Saha, Laurent Perrin, Laurence Röder, Christine Brun, Lionel Spinelli. Nucleic Acids Research, Volume 50, Issue 19, 28 October 2022, Page e114, https://doi.org/10.1093/nar/gkac715
- Genetic architecture of natural variation of cardiac performance from flies to humans. Saswati Saha, Lionel Spinelli, Laurence Roder, Christine Brun, Jaime A Castro Mondragon, et al. eLife 11:e82459. https://doi.org/10.7554/eLife.82459
- MYC and MET cooperatively drive hepatocellular carcinoma with distinct molecular traits and vulnerabilities. Sequera C, Grattarola M, Holczbauer A, Dono R, Pizzimenti S, Barrera G, Wangensteen KJ, Maina F. Cell Death Dis 13, 994 (2022). https://doi.org/10.1038/s41419-022-05411-6
- Universal multilayer network exploration by random walk with restart. Baptista, A., Gonzalez, A. & Baudot, A. . Commun Phys 5, 170 (2022). https://doi.org/10.1038/s42005-022-00937-9
- A Rare Mutation in LMNB2 Associated with Lipodystrophy Drives Premature Cell Senescence. Varlet, A.-A.; Desgrouas, C.; Jebane, C.; Bonello-Palot, N.; Bourgeois, P.; Levy, N.; Helfer, E.; Dubois, N.; Valero, R.; Badens, C.; et al. A Rare Mutation in LMNB2 Associated with Lipodystrophy Drives Premature Cell Senescence. Cells 2022, 11, 50. https://doi.org/10.3390/cells11010050
- Stoichiometry controls the dynamics of liquid condensates of associative proteins. Pierre Ronceray, Yaojun Zhang, Xichong Liu and Ned S. Wingreen, Phys. Rev. Lett. 128, 038102 (2022). arXiv:2101.0116
- Contrasting Roles of DOP as a Source of Phosphorus and Energy for Marine Diazotrophs, Filella Alba, Riemann Lasse, Van Wambeke France, Pulido-Villena Elvira, Vogts Angela, Bonnet Sophie, Grosso Olivier, Diaz Julia M., Duhamel Solange, Benavides Mar, Frontiers in Marine Science, Vol. 9 (2022), https://www.frontiersin.org/articles/10.3389/fmars.2022.923765, DOI: 10.3389/fmars.2022.923765, ISSN: 2296-7745
- Sinking Trichodesmium fixes nitrogen in the dark ocean. Benavides, M., Bonnet, S., Le Moigne, F.A.C. et al. ISME J 16, 2398–2405 (2022). https://doi.org/10.1038/s41396-022-01289-6
- Diazotrophs are overlooked contributors to carbon and nitrogen export to the deep ocean. Bonnet, S., Benavides, M., Le Moigne, F.A.C. et al. ISME J 17, 47–58 (2023). https://doi.org/10.1038/s41396-022-01319-3
- Planktonic Aggregates as Hotspots for Heterotrophic Diazotrophy: The Plot Thickens. Riemann Lasse, Rahav Eyal, Passow Uta, Grossart Hans-Peter, de Beer Dirk, Klawonn Isabell, Eichner Meri, Benavides Mar, Bar-Zeev Edo, Frontiers in Microbiology, Vol. 13 (2022), https://www.frontiersin.org/articles/10.3389/fmicb.2022.875050, DOI: 10.3389/fmicb.2022.875050, ISSN=1664-302X
- Lrrcc1 and Ccdc61 are conserved effectors of multiciliated cell function. Nommick, A., Boutin, C., Rosnet, O., Schirmer, C., Bazellieres, E., Thome, V., Loiseau, E., Viallat, A. and Kodjabachian, L. 2022. J. Cell Sci. 135(4):jcs258960.
- C. elegans: out on an evolutionary limb. (revue). Pujol N, Ewbank JJ.Immunogenetics. 2022 Feb;74(1):63-73. doi: 10.1007/s00251-021-01231-8. PMID: 34761293
2021
- Leukocyte transmigration and longitudinal forward-thrusting force in a microfluidic Transwell device. Aoun L, Nègre P, Gonsales C, Seveau de Noray V, Brustlein S, Biarnes-Pelicot M, Valignat MP, Theodoly O. Biophys J. 2021 Jun 1;120(11):2205-2221. doi: 10.1016/j.bpj.2021.03.037. Epub 2021 Apr 8.
- Functional Mapping of Adhesiveness on Live Cells Reveals How Guidance Phenotypes Can Emerge From Complex Spatiotemporal Integrin Regulation. Front Bioeng BiotechnolRobert P, Biarnes-Pelicot M, Garcia-Seyda N, Hatoum P, Touchard D, Brustlein S, Nicolas P, Malissen B, Valignat MP, Theodoly O. Front Bioeng Biotechnol. 2021 Apr 7;9:625366. doi: 10.3389/fbioe.2021.625366.
- Integrin-Functionalised Giant Unilamellar Vesicles via Gel-Assisted Formation: Good Practices and Pitfalls. Souissi, M.; Pernier, J.; Rossier, O.; Giannone, G.; Le Clainche, C.; Helfer, E.; Sengupta, K. Integrin-Functionalised Giant Unilamellar Vesicles via Gel-Assisted Formation: Good Practices and Pitfalls. Int. J. Mol. Sci. 2021, 22, 6335. https://doi.org/10.3390/ijms22126335
- Nucleation landscape of biomolecular condensates. Shimobayashi, S.F., Ronceray, P., Sanders, D.W. et al. Nature (2021). DOI: 10.1038/s41586-021-03905-5
- Self-organization and shape change by active polarization in nematic droplets, Fabian Jan Schwarzendahl, Pierre Ronceray, Kimberly L. Weirich, and Kinjal Dasbiswas, Phys. Rev. Research 3, 043061 (2021). arXiv:2102.07442
- Physical bioenergetics: Energy fluxes, budgets, and constraints in cells, X Yang, M Heinemann, J Howard, G Huber, S Iyer-Biswas, G Le Treut, M Lynch, KL Montooth, DJ Needleman, S Pigolotti, J Rodenfels, P Ronceray, S Shankar, I Tavassoly, S Thutupalli, DV Titov, J Wang, PJ Foster,
- Misic, a general deep learning-based method for the high-throughput cell segmentation of complex bacterial communities. Swapnesh Panigrahi, Dorothée Murat, Antoine Le Gall, Eugénie Martineau, Kelly Goldlust, Jean-Bernard Fiche, Sara Rombouts, Marcelo Nöllmann, Leon Espinosa, Tâm Mignot. eLife. 2021 Sept. 9;10:e65151. DOI: 10.7554/eLife.65151.
- NF-κB-dependent IRF1 activation programs cDC1 dendritic cells to drive antitumor immunity. Ghislat G, Cheema AS, Baudoin E, Verthuy C, Ballester PJ, Crozat K, Attaf N, Dong C, Milpied P, Malissen B, Auphan-Anezin N, Manh TPV, Dalod M, Lawrence T. Sci Immunol. 2021 Jul 9;6(61):eabg3570. doi: 10.1126/sciimmunol.abg3570. PMID: 34244313
- Evolution of mechanisms controlling epithelial morphogenesis across animals: new insights from dissociation-reaggregation experiments in the sponge Oscarella lobularis. Vernale A, Prünster MM, Marchianò F, Debost H, Brouilly N, Rocher C, Massey-Harroche D, Renard E, Le Bivic A, Habermann BH, Borchiellini C BMC Ecol Evo 21, 160 (2021). doi: 10.1186/s12862-021-01866-x. PMID: 34418961.
- Complete Genome Assembly of Myxococcus xanthus Strain DZ2 Using Long High-Fidelity (HiFi) Reads Generated with PacBio Technology. Jain R, Habermann BH, Mignot T. Microbiol Resour Announc. 2021 Jul 15;10(28):e0053021. doi: 10.1128/MRA.00530-21. Epub 2021 Jul 15. PMID: 34264106; PMCID: PMC8281077.
- . STEM CELLS Transl Med. 2021; 10: 725– 742. https://doi.org/10.1002/sctm.20-0177
- MetamORF: a repository of unique short open reading frames identified by both experimental and computational approaches for gene and metagene analyses, Database. Sebastien A Choteau, Audrey Wagner, Philippe Pierre, Lionel Spinelli, Christine Brun. Volume 2021, 2021, baab032, https://doi.org/10.1093/database/baab032
- Formation of polarized contractile interfaces by self-organized Toll-8/Cirl GPCR asymmetry. Lavalou J, Mao Q, Harmansa S, Kerridge S, Lellouch AC, Philippe JM, Audebert S, Camoin L, Lecuit T. Dev Cell. 2021 Apr 27:S1534-5807(21)00270-7. doi: 10.1016/j.devcel.2021.03.030. Online ahead of print. PMID: 33932333
- Distinct actin-dependent nanoscale assemblies underlie the dynamic and hierarchical organization of E-cadherin. Chandran R, Kale G, Philippe JM, Lecuit T, Mayor S. Curr Biol. 2021 Apr 26;31(8):1726-1736.e4. doi: 10.1016/j.cub.2021.01.059. Epub 2021 Feb 18. PMID: 33607036
- Programmed and self-organized flow of information during morphogenesis. Collinet C, Lecuit T. Nat Rev Mol Cell Biol. 2021 Apr;22(4):245-265. doi: 10.1038/s41580-020-00318-6. Epub 2021 Jan 22. PMID: 33483696 Review.
2020
- Spot Variation Fluorescence Correlation Spectroscopy for Analysis of Molecular Diffusion at the Plasma Membrane of Living Cells. Mailfert S, Wojtowicz K, Brustlein S, Blaszczak E, Bertaux N, Łukaszewicz M, Marguet D, Trombik T. J Vis Exp. 2020 Nov 12;(165). doi: 10.3791/61823.
- Neuronal specification in elegans: combining lineage inheritance with intercellular signaling. Barrière A. and Bertrand V. (2020). J. Neurogenetics 34, 273-81. DOI : 10.1080/01677063.2020.1781850.
- Wnt ligands regulate the asymmetric divisions of neuronal progenitors in elegans embryos. Kaur S., Mélénec P., Murgan S., Bordet G., Recouvreux P., Lenne P.F. and Bertrand V. (2020). Development 147, dev183186.
- Long-read only assembly of Drechmeria coniospora genomes reveals widespread chromosome plasticity and illustrates the limitations of current nanopore methods. Courtine D, Provaznik J, Reboul J, Blanc G, Benes V, Ewbank JJ.. Gigascience. 2020 Sep 18;9(9):giaa099. doi: 10.1093/gigascience/giaa099.
- Non-Canonical Caspase Activity Antagonizes p38 MAPK Stress-Priming Function to Support Development. Weaver BP, Weaver YM, Omi S, Yuan W, Ewbank JJ, Han MDev Cell. 2020 May 4;53(3):358-369.e6. doi: 10.1016/j.devcel.2020.03.015. Epub 2020 Apr 16. PMID: 32302544.
- Fabrication of sharp silicon arrays to wound Caenorhabditis elegans. Belougne J, Ozerov I, Caillard C, Bedu F, Ewbank JJ. Sci Rep. 2020 Feb 27;10(1):3581. doi: 10.1038/s41598-020-60333-7. PMID: 32108170.
- Microtubule plus-end dynamics link wound repair to the innate immune response. Taffoni C, Omi S, Huber C, Mailfert S, Fallet M, Rupprecht JF, Ewbank JJ, Pujol N. Elife. 2020 Jan29;9:e45047. doi: 10.7554/eLife.45047. PMID: 31995031.
- New Strains for Tissue-Specific RNAi Studies in Caenorhabditis elegans. Watts JS, Harrison HF, Omi S, Guenthers Q, Dalelio J, Pujol N, Watts JL. G3 (Bethesda). 2020 Nov 5;10(11):4167-4176. doi: 10.1534/g3.120.401749. PMID: 32943454.
- IL-17: good fear no tears. Rua R, Pujol N. Nat Immunol. 2020 Nov;21(11):1315-1316. doi: 10.1038/s41590-020-0792-4. PMID: 32958929.
- Diversity from Similarity: Cellular Strategies for Assigning Particular Identities to Actin Filaments and Networks. Boiero Sanders M., Antkowiak A., Michelot A*. Open Biol. 2020 Sep;10(9):200157. doi: 10.1098/rsob.200157.
- Force Production by a Bundle of Growing Actin Filaments is Limited by its Mechanical Properties. Martiel J.L., Michelot Alphée, Boujemaa-Paterski R., Blanchoin L., Berro J.. Biophys J. 2020 Jan 7;118(1):182-192. doi: 10.1016/j.bpj.2019.10.039.
- Assembly of a persistent apical actin network by the formin Frl/Fmnl tunes epithelial cell deformability. Dehapiot, B., Clément, R., Alégot, H. et al. Nat Cell Biol (2020). https://doi.org/10.1038/s41556-020-0524-x.
- Coopted temporal patterning governs cellular hierarchy, heterogeneity and metabolism in Drosophila neuroblast tumors. Genovese S, Clément R, Gaultier C, Besse F, Narbonne-Reveau K, Daian F, Foppolo S, Luis NM, Maurange Elife. 2019 Sep 30;8:e50375. doi: 10.7554/eLife.50375. PMID: 31566561.
- Towards a general framework for spatio-temporal transcriptomics – LMRL Workshop, Neurips 2020 - J. Pinol, T. Artières, P. Villoutreix. Youtube link.
- Human neutrophils swim and phagocytise bacteria. Garcia-Seyda N, Seveau V, Manca F, Biarnes-Pelicot M, Valignat M-P, Bajénoff M and Theodoly O*. Biology of the Cell, doi: 10.1111/boc.202000084.
- Amoeboid Swimming Is Propelled by Molecular Paddling in Lymphocytes. Aoun L; Farutin, A; Garcia-Seyda, N; Negre, P; Rizvi, MS; Tlili, S (Tlili, Sham)[ 1,4 ] ; Song, S (Song, Solene)[ 1 ] ; Luo, X; Biarnes-Pelicot, M; Galland, R; Sibarita, JB ; Michelot; Hivroz, C; Rafai, S; Valignat, MP ; Misbah*, C; Theodoly, O*, Biophysical Journal, 119, 6,1157-1177, doi: 10.1016/j.bpj.2020.07.033.
- Lymphocytes perform reverse adhesive haptotaxis mediated by LFA-1 integrins. Luo, X ; de Noray, VS ; Aoun, L; Biarnes-Pelicot, M ; Strale, PO; Studer, V; Valignat, MP; Theodoly, O*. Journal of Cell Science, 133, 16, Article Number: jcs242883, DOI: 10.1242/jcs.242883, 2020.
- Three-dimensional imaging with reflection synthetic confocal microscopy. Rasedujjaman, M ; Affannoukou, K; Garcia-Seyda, N; Robert, P; Giovannini, H ; Chaumet, PC; Theodoly O; Valignat, MP; Belkebir,; Sentenac, A; Guillaume Maire . Optics Letters 2020, 45, 13, 3721-3724. doi: 10.1364/OL.397364.
- Microfluidic device to study flow-free chemotaxis of swimming cells. Garcia-Seyda N, Aoun L, Tishkova V, Seveau V, Biarnes-Pelicot M, Bajenoff M, Valignat MP, Olivier Theodoly* Chip. 2020;20(9):1639–1647, (2020). doi: 10.1039/d0lc00045k.
- A Bistable Mechanism Mediated by Integrins Controls Mechanotaxis of Leukocytes. A. Hornung, T. Sbarrato, N. Garcia-Seyda, L. Aoun, X. Luo, M. Biarnes-Pelicot, Theodoly*, MP. Valignat*. Biophys. J. 2020;118(3):565–577, (2020). DOI: 10.1016/j.bpj.2019.12.013.
- Deterministic and Stochastic Rules of Branching Govern Dendrite Morphogenesis of Sensory Neurons. Palavalli A, Tizón-Escamilla N, Rupprecht JF, Lecuit T. Curr Biol. 2021 Feb 8;31(3):459-472.e4. doi: 10.1016/j.cub.2020.10.054. Epub 2020 Nov 18.
- Adhesion-mediated heterogeneous actin organization governs apoptotic cell extrusion. A. P. Le, J.-F. Rupprecht, R.-M. Mège, Y. Toyama, C. T. Lim, B. Ladoux, Nature Communications (2020).
- Embryonic geometry underlies phenotypic variation in decanalized conditions. A. Huang, J.-F. Rupprecht and T. E. Saunders eLife (2020).
- Measuring social networks in primates: wearable sensors vs. direct observations, V. Gelardi, J. Godard, D. Paleressompoulle, N. Claidière, A. Barrat, Proc. R. Soc. A 476:20190737 (2020)
- Relevance of temporal cores for epidemic spread in temporal networks, M. Ciaperoni, E. Galimberti, F. Bonchi, C. Cattuto, F. Gullo, A. Barrat, Scientific Reports 10:12529 (2020)
- Dynamic core periphery structure of information sharing networks in entorhinal cortex and hippocampus, N. Pedreschi, C. Bernard, W. Clawson, P. Quilichini, A. Barrat, D. Battaglia, Network Neuroscience 4:946 (2020)
- Modeling Heterogeneity of Triple-Negative Breast Cancer Uncovers a Novel Combinatorial Treatment Overcoming Primary Drug Resistance. Lamballe F., Ahmad F., Vinik Y., Castellanet O., Daian F., Müller A.K., Köhler U.A., Bailly A.L., Josselin E., Castellano R. Cayrou C., Charafe-Jauffret E., Mills G.B., Géli V., Borg J.P., Lev S., Maina F. Advanced Science, Dec 16;8(3):2003049 (2020). PMID: 33552868.
- Enhanced differentiation of human induced pluripotent stem cells towards the midbrain dopaminergic neuron lineage through GLYPICAN-4 down regulation. Corti S., Bonjean R., Rattier D., Legier T., Melon C., Salin P., Toso E.A., Kyba M., Kerkerian-Le Goff L., Maina F., Dono R. Stem Cells Translational Medicine, Feb 2 (2021). PMID: 33528918.
- C3G is upregulated in hepatocarcinoma, contributing to tumor growth and progression and to HGF/MET pathway activation. Sequera C., Bragado P., Manzano S., Arechederra M., Richelme S., Gutiérrez-Uzquiza A., Sánchez A., Maina F., Guerrero C., Porras A. Cancers, Aug 14;12(8):2282 (2020). PMID: 32823931.
- The Dorsal Striatum Energizes Motor Routines. Jurado-Parras MT, Safaie M, Sarno S, Louis J, Karoutchi C, Berret B, Robbe D. Current biology : CB - Sep 2020
- Turning the body into a clock: Accurate timing is facilitated by simple stereotyped interactions with the environment. , , , , , , , , and
- The activation trajectory of plasmacytoid dendritic cells in vivo during a viral infection. Abbas A, Vu Manh TP, Valente M, Collinet N, Attaf N, Dong C, Naciri K, Chelbi R, Brelurut G, Cervera-Marzal I, Rauwel B, Davignon JL, Bessou G, Thomas-Chollier M, Thieffry D, Villani AC, Milpied P, Dalod M, Tomasello E. Nat Immunol. 2020. doi: 10.1038/s41590-020-0731-4.
- The quest for faithful in vitro models of human dendritic cells types. Luo XL, Dalod M. Mol Immunol. 2020. doi: 10.1016/j.molimm.2020.04.018.
- Anisotropy links cell shapes to tissue flow during convergent extension. Xun Wang, Matthias Merkel, Leo B. Sutter, Gonca Erdemci-Tandogan, M. Lisa Manning, and Karen E. Kasza. PNAS June 16, 2020 117 (24) 13541-13551; first published May 28, 2020; https://doi.org/10.1073/pnas.1916418117.
- An NF-κB/IRF1 axis programs cDC1s to drive anti-tumor immunity. Ghislat G, Cheema AS, Baudoin E, Verthuy C, Ballester PJ, Crozat K, Attaf N, Dong C, Milpied P, Malissen B, Auphan-Anezin N, Vu Manh TP, Dalod M, Lawrence T. bioRxiv 2020.12.28.424621; doi: https://doi.org/10.1101/2020.12.28.424621.
- Active mucus–cilia hydrodynamic coupling drives self-organization of human bronchial epithelium.Loiseau, E., Gsell, S., Nommick, A. et al. Nat. Phys. (2020). https://doi.org/10.1038/s41567-020-0980-z.
- NK cells orchestrate splenic cDC1 migration to potentiate antiviral protective CD8+ T cell responses. Ghilas S, Ambrosini M, Cancel JC, Massé M, Lelouard H, Dalod M, Crozat K. bioRxiv 2020.04.23.057463; doi: https://doi.org/10.1101/2020.04.23.057463.
- cDC1 and interferons promote spontaneous CD4+ and CD8+ T cell protective responses to breast cancer. Mattiuz R, Brousse C, Ambrosini M, Cancel JC, Bessou G, Mussard J, Sanlaville A, Caux C, Bendriss-Vermare N, Valladeau-Guilemond J, Dalod M, Crozat K. bioRxiv 2020.12.23.424092; doi: https://doi.org/10.1101/2020.12.23.424092
- Networks beyond pairwise interactions: Structure and dynamics. Federico Battiston, Giulia Cencetti, Iacopo Iacopini, Vito Latora, Maxime Lucas, Alice Patania, Jean-Gabriel Young, and Giovanni Petri. Physics Reports, 874:1--92, 2020. Networks beyond pairwise interactions: Structure and dynamics.
- Hippocampal hub neurons maintain distinct connectivity throughout their lifetime. Bocchio M, Gouny C, Angulo-Garcia D, Toulat T, Tressard T, Quiroli E, Baude A, Cossart R. Nat Commun. 2020 Sep 11;11(1):4559. doi: 10.1038/s41467-020-18432-6. PMID: 32917906 Free PMC article.
- Antagonistic fungal enterotoxins intersect at multiple levels with host innate immune defences. Xing Zhang, Benjamin Harding, Dina Aggad, Damien Courtine, Jia-Xuan Chen, Nathalie Pujol, and Jonathan Ewbank.
- Multiorder laplacian for synchronization in higher-order networks. Maxime Lucas, Giulia Cencetti, and Federico Battiston. Phys. Rev. Research, 2:033410, Sep 2020.
- DeepCINAC: A Deep-Learning-Based Python Toolbox for Inferring Calcium Imaging Neuronal Activity Based on Movie Visualization. Denis J, Dard RF, Quiroli E, Cossart R, Picardo MA. eNeuro. 2020 Aug 17;7(4):ENEURO.0038-20.2020. doi: 10.1523/ENEURO.0038-20.2020. Print 2020 Jul/Aug. PMID: 32699072.
- GABAergic Restriction of Network Dynamics Regulates Interneuron Survival in the Developing Cortex. Duan ZRS, Che A, Chu P, Modol L, Bollmann Y, Babij R, Fetcho RN, Otsuka T, Fuccillo MV, Liston C, Pisapia DJ, Cossart R, De Marco García NV. Neuron. 2020 Jan 8;105(1):75-92.e5. doi: 10.1016/j.neuron.2019.10.008. Epub 2019 Nov 25. PMID: 31780329 Free PMC article.
- Assemblies of Perisomatic GABAergic Neurons in the Developing Barrel Cortex. Modol L, Bollmann Y, Tressard T, Baude A, Che A, Duan ZRS, Babij R, De Marco García NV, Cossart R. Neuron. 2020 Jan 8;105(1):93-105.e4. doi: 10.1016/j.neuron.2019.10.007. Epub 2019 Nov 25. PMID: 31780328 Free PMC article.
2019
- Self-organization of red blood cell suspensions under confined 2D flows. C. Iss, D. Midou, A. Moreau, D. Held, A. Charrier, S. Mendez, A. Viallat, E. Helfer. Soft Matter 15, 2971 (2019)
- White blood cells dynamics in micro-flows. A. Viallat, J. Dupire, E. Helfer. In : Blood Cells Suspension Dynamics in Microflows, CRC Press (2019)
- Are Conventional Type 1 Dendritic Cells Critical for Protective Antitumor Immunity and How? Cancel JC, Crozat K, Dalod M, Mattiuz R. Front Immunol. 2019;10:9. doi: 10.3389/fimmu.2019.00009.
- A minimal-length approach unifies rigidity in underconstrained materials. Matthias Merkel, Karsten Baumgarten, Brian P. Tighe, M. Lisa Manning. Proceedings of the National Academy of Sciences Apr 2019, 116 (14) 6560-6568; DOI: 10.1073/pnas.1815436116
- Matrix metalloproteinase 1 modulates invasive behavior of tracheal branches during entry into Drosophila flight muscles. Sauerwald J, Backer W, Matzat T, Schnorrer F, Luschnig S. eLife. pii: e48857. doi: 10.7554/eLife.48857.
- A small proportion of Talin molecules transmit forces at developing muscle attachments in vivo. Lemke SB, Weidemann T, Cost AL, Grashoff C, and Schnorrer F. (2019). PLoS Biology. 17(3):e3000057. doi: 10.1371/journal.pbio.3000057
- 1 contributes to a rapid homeostatic plasticity of intrinsic excitability in CA1 pyramidal neurons in vivo. Morgan PJ, Bourboulou R, Filippi C, Koenig-Gambini J, Epsztein J. Elife. 2019 Nov 27;8.
- Dynamic control of hippocampal spatial coding resolution by local visual cues. Bourboulou R, Marti G, Michon FX, El Feghaly E, Nouguier M, Robbe D, Koenig J, Epsztein J. Elife. 2019 Mar 1;8
- Genetic induction and mechanochemical propagation of a morphogenetic wave; Baille, A, Collinet C, Philippe JM, Lenne PF, Munro E, Lecuit T. Nature. 2019 Aug;572(7770):467-473. doi: 10.1038/s41586-019-1492-9. Epub 2019 Aug 15. PMID: 31413363
- Experimental validation of force inference in epithelia from cell to tissue scale; Kong W, et al. Sci Rep 2019. PMID 31601854
- Mechanical Stiffness of Reconstituted Actin Patches Correlates Tightly with Endocytosis Efficiency. PLANADE J, BELBAHRI R, BOEIRO SANDERS M, GUILLOTIN A, DU ROURE O, MICHELOT A*, HEUVINGH J. PLoS Biol. 2019 Oct 25 ;17(10) :e3000500
- Sizes of actin networks sharing a common environment are determined by the relative rates of assembly. ANTKOWIAK A, GUILLOTIN A, BOEIRO SANDERS M, COLOMBO J, VINCENTELLI R, MICHELOT A*. PLoS Biol. 2019 Jun 10 ;17(6) :e3000317
- Internal representation of hippocampal neuronal population spans a time-distance continuum. Haimerl C, Angulo-Garcia D, Villette V, Reichinnek S, Torcini A, Cossart R, Malvache A. Proc Natl Acad Sci U S A. 2019 Apr 9;116(15):7477-7482. doi: 10.1073/pnas.1718518116
- Distinct RhoGEFs activate apical and junctional actomyosin contractility under control of G proteins during epithelial morphogenesis. Alain Garcia De Las Bayonas, Jean-Marc Philippe, Annemarie C. Lellouch, Thomas Lecuit. Current Biology. 2019 Oct 21;29(20):3370-3385.e7. DOI: 10.1016/j.cub.2019.08.017
2018
- Distinct contributions of tensile and shear stress on E-cadherin levels during morphogenesis.Kale GR, Yang X, Philippe JM, Mani M, Lenne PF, Lecuit T. Nat Commun. 2018 Nov 27;9(1):5021. doi: 10.1038/s41467-018-07448-8. PMID: 30479400
- Novel Cre-Expressing Mouse Strains Permitting to Selectively Track and Edit Type 1 Conventional Dendritic Cells Facilitate Disentangling Their Complexity in vivo. Mattiuz R, Wohn C, Ghilas S, Ambrosini M, Alexandre YO, Sanchez C, Fries A, Vu Manh TP, Malissen B, Dalod M, Crozat K. Front Immunol. 2018;9:2805. doi: 10.3389/fimmu.2018.02805.
- Quantitative Control of GPCR Organization and Signaling by Endocytosis in Epithelial Morphogenesis. Jha A, van Zanten TS, Philippe JM, Mayor S, Lecuit T., Curr Biol. 2018 May 21;28(10):1570-1584.e6. doi: 10.1016/j.cub.2018.03.068. Epub 2018 May 3.
- Strengthen Your Junctions to Resist the Force. Lellouch AC, de Las Bayonas AG, Lecuit T. Dev Cell. 2018 Nov 19;47(4):406-407. doi: 10.1016/j.devcel.2018.11.007. PMID: 30458136