CENTURI Hackathon
2024
CENTURI Hackathon
2024
For quantitative biology
Centre de séminaires
Villages Clubs du Soleil, Belle de Mai, Marseille
What is the CENTURI Hackathon?
The CENTURI Hackathon is a fast-paced two-day event of coding, engineering and idea-sharing to drive innovations at the interface of Computer Science and Life Sciences. The event is aimed at curious and creative minds interested in improving their coding skills through collaborative projects. The event is open to students, postdocs, developers and researchers.
The goal of our Hackathon is to computationally unlock technological bottlenecks arising from the study of Living Systems. Our event is aimed at everyone with experience in coding and with a keen interest in problem solving in big data, computer vision or modeling. Experts in fields such as biology, mathematics and non-scientific developers are encouraged to apply!
Curious about the previous editions? Find out more about the 2022 Hackathon, the 2023 Hackathon and the 2023 Hackathon projects here!

What types of projects will participants be working on ?
Projects will strive to solve key technological bottlenecks in a variety of topics, such as Computer vision, Modeling, Data mining and Computer assisted microscopy.
Participants will work on big data projects for living systems, with projects from academic labs in CENTURI and elsewhere.
CENTURI Hackathon will welcome a maximum of 60 participants. Applicants should have some experience with coding.
Participants will work in groups of 6-8 people on various projects in a variety of topics:
- Data visualization and interactions
- Computer vision
- Data mining
- Robotics & Computer assisted microscopy
- Modelling
Projects are as follows:
EmbryoFlowTracker | Léo Guignard
Keywords: Embryo Development, Tracking Algorithms, Vector Flow
Understanding the formation of life from a single fertilised cell requires a detailed description of this process. Many researchers have focused on deciphering cell dynamics, studying how cells organised into tissues, organs, and eventually an entire organism.
Considerable progress has been made in quantifying cell movement within whole embryos. This progress began with advancements in microscope technologies, such as light-sheet microscopy, allowing researchers to observe all the cells in an embryo over extended periods.
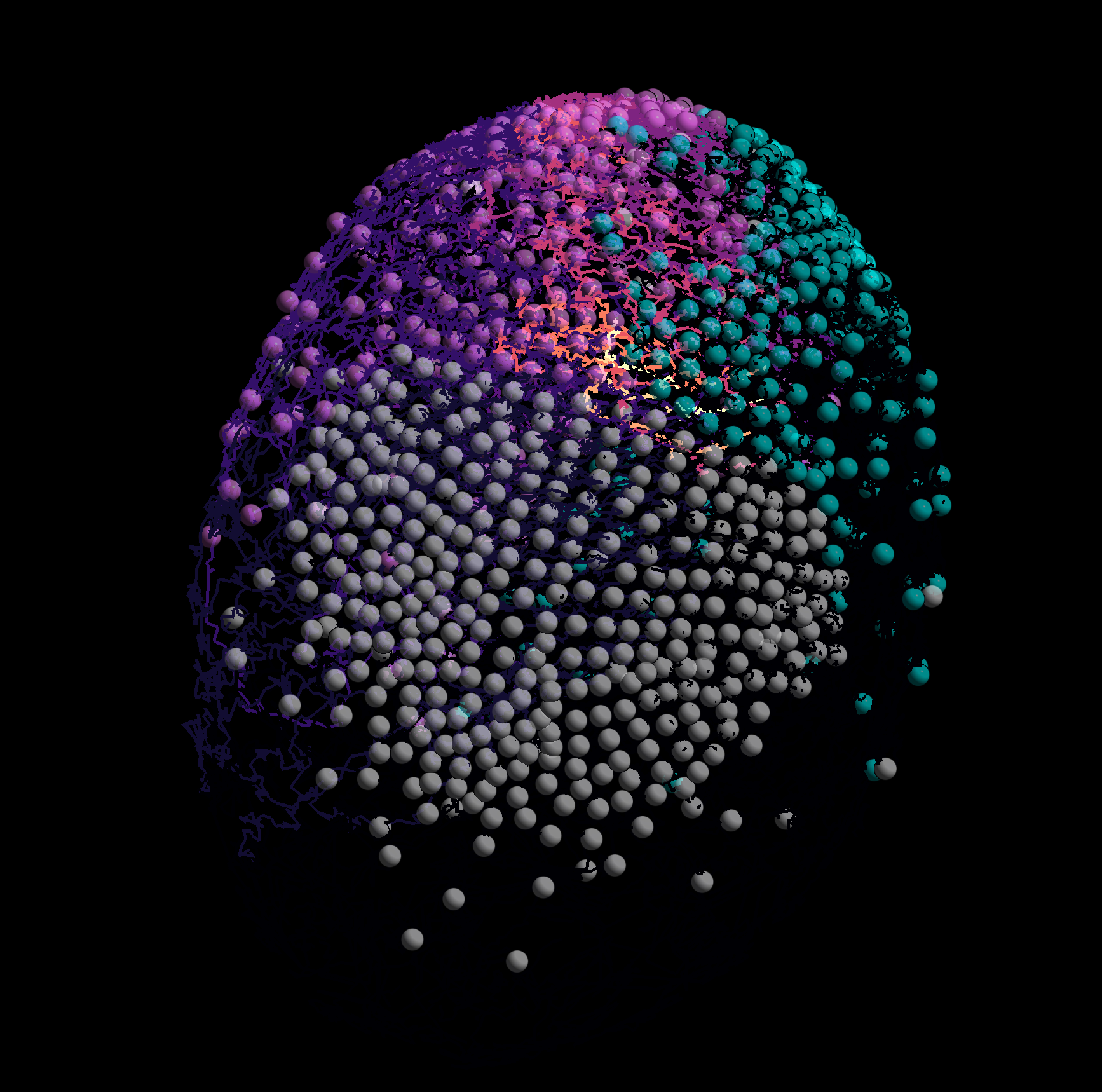
However, extracting quantitative data from these complex datasets required the development of specialised computational tools.
Despite advancements, challenges remain in automating cell identification and tracking within 3D movies. In this project, the team will work with large 3D+time datasets to improve tracking algorithms using precise non-linear vector fields.
To support algorithm development, the team will have access to resources including 500 time points of a 3D movie showing the development of a Parhyale embryo, loose segmentation data for each time point, and non-linear vector fields linking consecutive time points. Additionally, to validate their method, they will have manual partial cell detection and tracking data for over 400 time points.
This interdisciplinary effort aims to shed light on the intricate processes of cell behaviour during embryo development, using advanced technology and analytical approaches.
Technical and Technological Bottlenecks: The main technical bottleneck lies in developing efficient algorithms to accurately track cells using non-linear vector fields. Current approaches lack the precision required for this task.
Programming Language & Toolbox: The algorithm will be implemented in Python, leveraging libraries such as NumPy, SciPy, and scikit-learn for data processing and machine learning. Visualisation will be handled with Matplotlib and Plotly.
Expected Deliverable / Output: The team aims to deliver "EmbryoFlowTracker," a software tool for precise cell tracking in developing embryos using non-linear vector fields. This tool will provide researchers with a user-friendly interface to analyse 3D+time datasets of embryo development. Additionally, the project will produce a research paper detailing the algorithm's performance and its application to the provided Parhyale embryo dataset.
DeepForce: Deep single molecule unfolding detection | Felix Rico, Claire Valotteau, Ismahene Mesbah
Keywords: Force Spectroscopy, Unfolding, Event Detection, Neural Networks
Receptor-ligand bonds are a central part of any biological process and protein folding is one of the unsolved problems in biology. These two processes can be probed at the single molecule level using force spectroscopy experiments
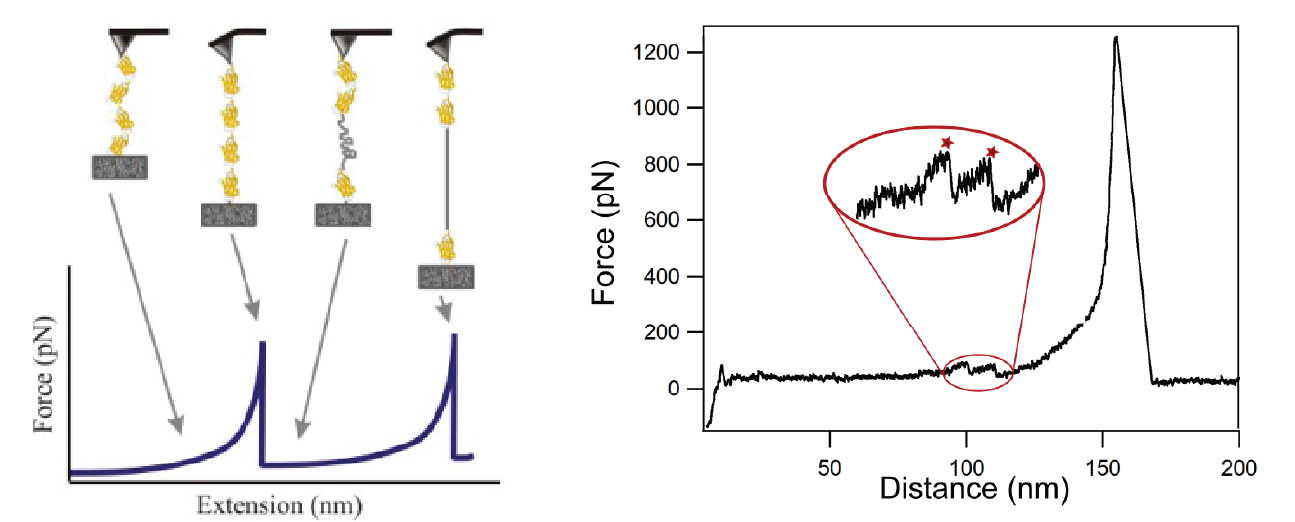
Single Molecule Force Spectroscopy is based on the acquisition of force-distance curves in which a single molecule, e.g. a protein, is pulled and force required to unfold or unbind it is measured. Experimentally, only about 10% of these curves show an interesting signature (unbinding and/or unfolding peak), the others show no event. An example of the output is given in the figure below. The automatic detection of those noisy and heterogeneous events is very challenging and requires specific tools for an accurate and trustworthy analysis of molecular mechanics.
For this hackathon, we will develop sensitive deep-learning methods to pre-select the curves with interesting events, and then to classify the events and extract the molecular forces. We envision the following milestones to carry out this project.
- Training a recurrent neural network on simple simulation
- Experiment with real annotated data
- Development of a simple GUI for results exploration and re-training
Programming language & Toolbox Python, Matlab...
Dataset: Thousands of force curves from atomic force microscopy measurements of single molecule unfolding. Simulated force curves mimicking AFM experimental results.
EndoTrack: visualizing the movements of intracellular organelles in developing tissues | Claudio Collinet
Keywords: Morphogenesis, Visualization, Cell Dynamics, Multiple object tracking.
Morphogenesis is the emergence of new shapes in tissues and organisms during their embryonic development. Understanding this process is crucial to studying its disruption in embryonic malformations or tumor formation. Imaging experiments on embryos, such as those of the fruitfly Drosophila, uncovered that cells move, intercalate and change geometry to drive morphogenesis.
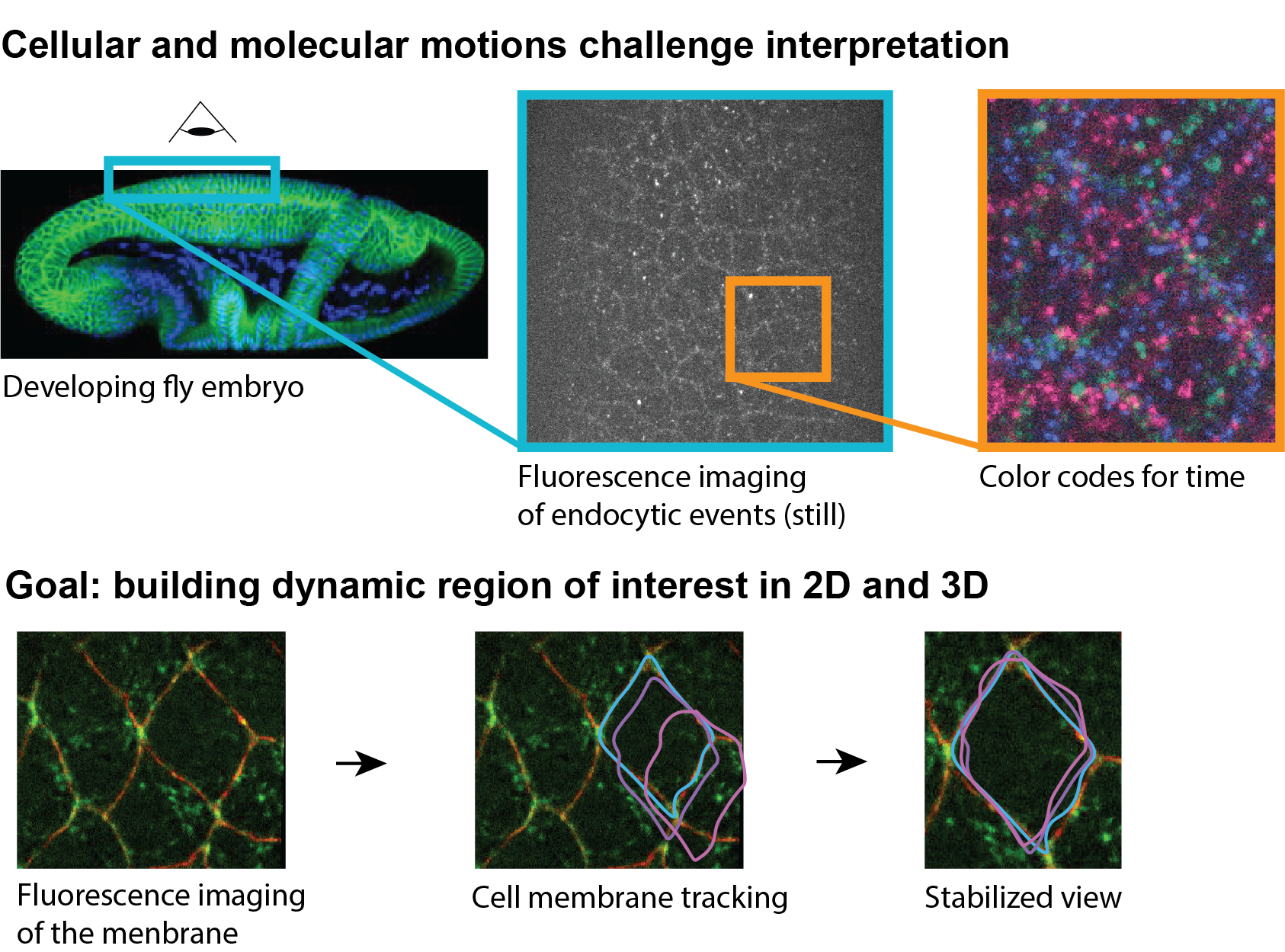
These processes are driven by fundamental intracellular processes, such as endocytic trafficking, where the collective dynamics of hundreds of intracellular organelles allow the redistribution of membrane molecules within the cell. Despite progress in 3D live imaging, the exact role of the endocytic process is poorly understood due to the complexity of the volumetric sequence. Indeed, visualization and interpretation are challenged by the multifactorial origins of motions (coming from cellular displacement, membrane reorganization and endocytosis itself) as well as its stochastic nature.
In this context, the goal of this project is the development of a toolset to change the ways we look at volumetric sequences. Instead of manually navigating a complex dataset, the user will be able to select a cell of interest to automatically visualize a dynamic 3D ROIs that follow the cell as they move through the sample. In those new movies devoid of cellular motion, the delicate orchestration of endocytic events will be now observable and even quantifiable. We have identified the following milestone toward this goal:
- Automatizing the tracking of cell on the epithelium in 2D first using membrane markers (Gap43 or Dextran) using off-the-shelf 2D segmentation tools (e.g. https://github.com/baigouy/EPySeg), tracking approach (e.g. https://github.com/DanuserLab/u-track) and 2D annotation for validation.
- Porting this implementation to 3D using the 2D pre-segmentation and motion as a starting point. Endocytic trackability (https://github.com/DanuserLab/u-track3D) could be used to evaluate the results.
- Build a dynamic ROI program that follows a single tracked cell and provides a static view.
Datasets: 3D time lapse images of Drosophila embryo acquired at IBDM by Claudio Collinet.
Programming Language & Toolbox: Python and optionally matlab
MetaGem: synthetic metabolic flux representation with generative AI. | Bianca Habermann
Keywords: Metabolism, Generative AI, Diffusion Model, Visualization.
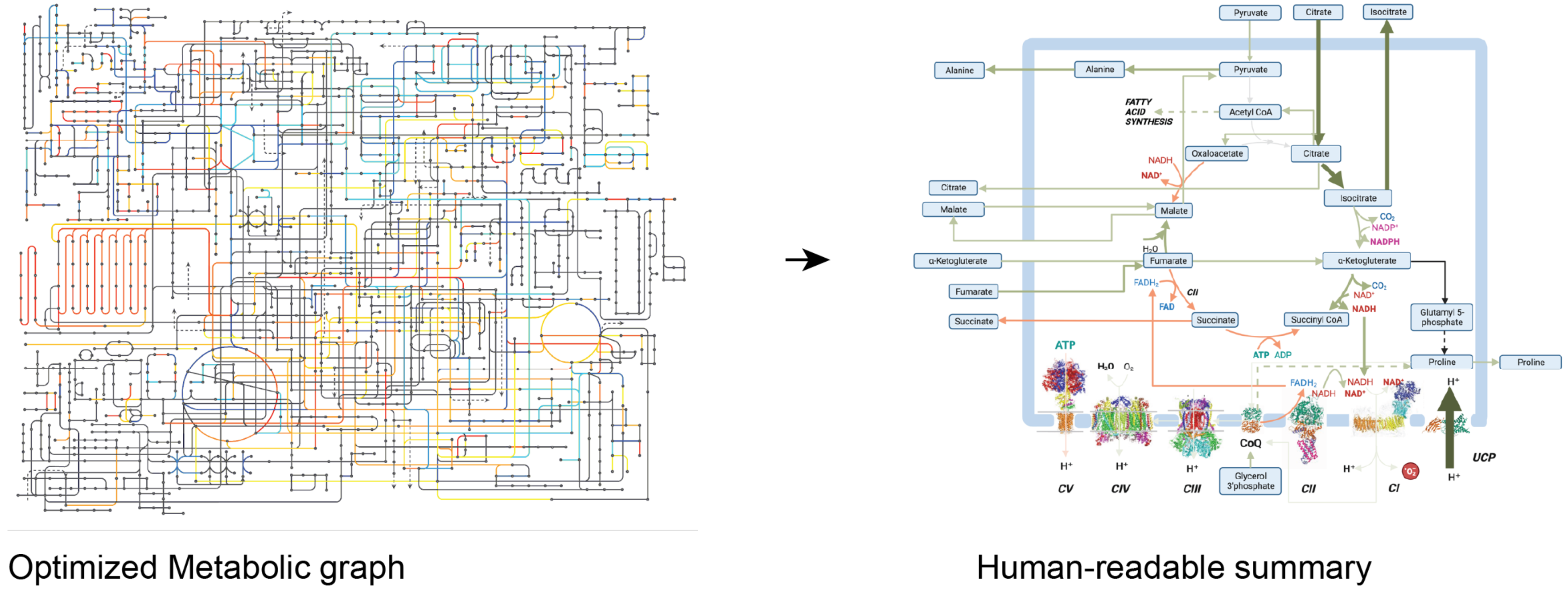
Metabolism is a cellular process that uses enzymes to provide and consume energy for movement, growth, development, and reproduction. Establishing the chain of metabolic interaction, a.k.a. metabolic flux is challenged by the lack of direct measurements as well as the cheer size of the metabolic network. To peer into this complex system, our interdisciplinary group develops new optimization methods to predict metabolite flux through the modeling of external stimuli, enzyme availability and interactions. This approach is called Flux Balance Analysis and maps out metabolite interaction in the whole graph. However, the resulting graph remains large, and representing the most pertinent interactions in a synthetic and interpretable manner is often a manual and labor intensive process.
Building upon recent breakthroughs in the field of stable diffusion to summarize complex sequential dataset and stable diffusion for image generation , this project will focus on the use of generative networks for the synthesis of human-readable illustration from the estimated graph of metabolic flux. We have devised the following preliminary milestones to help drive experimentations. However, the participants will be free to experiment with their tools of choice.
- The first step will focus on the training of a “text to image” generative model using formatted graph files and illustrative summary produced by experts. Stable diffusion approaches (e.g. https://github.com/CompVis/stable-diffusion) can be explored but must be adapted to the specific structure of metabolic graph.
- To test the quality of the resulting illustration, we will combine image-to-text, expert written descriptions and text distance metrics.
- An independent milestone will focus on experimentation with user interactions:
- How can a user customize or correct an illustration generated by the network.
- How can a user detect errors in the graph ?
- Try new interaction paradigm with complex graphs (3D view etc…)
- Enabling “zoom” on a generated graph for more details on the pathway.
- Automated “multiscale” decomposition to build coarse-to-fine representations that remain accessible to the naïve reader.
StemControlAI: Advancing Stem Cell Quality Control with Automation | Virgile Viasnoff
Keywords: Stem Cells, Robotic, AI, Cell Recognition
AI presents a compelling solution for streamlining the labour-intensive processes involved in cultivating high-quality stem cells.
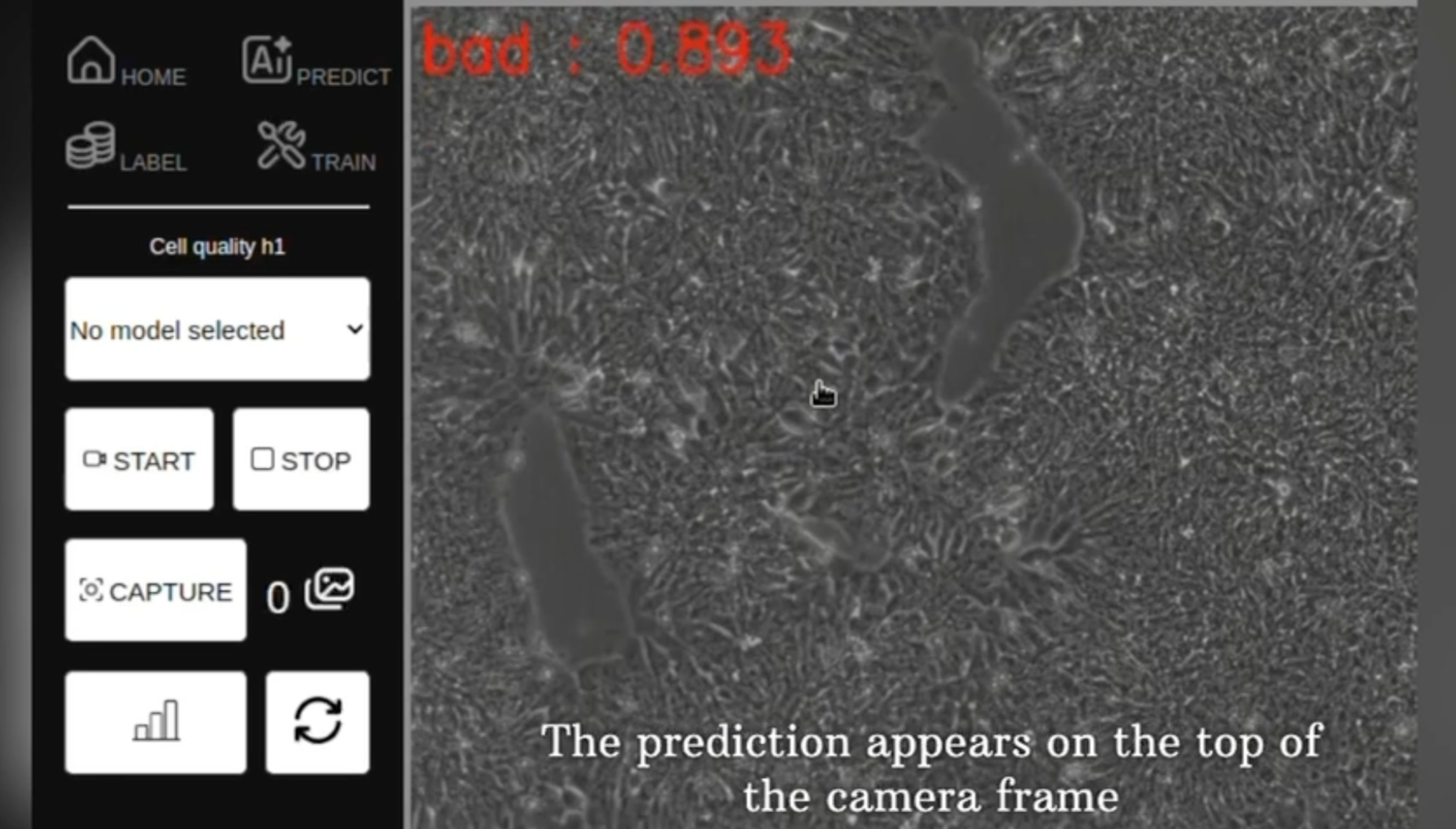
Within the industry, there's a growing imperative to integrate automated AI-based quality control into the assessment of stem cell and 3D culture quality. Currently, these assessments rely on human observation by skilled operators, lacking automation. They serve as critical checkpoints before engaging the next steps of differentiation protocols.
Our project aims to develop a smart camera system empowered by an AI-based recognition algorithm. This system will be seamlessly integrated with a custom-built microscope and robotic arm, engineered by the Mechanic Platform at the Turing Center. Raspberry Pi controllers are used to drive both the camera and the microscope, ensuring cost-effectiveness and accessibility.
The project's focal point is establishing the coding infrastructure to merge AI detection with the robotic arm, resulting in an affordable, automated system for AI-guided stem cell culture and differentiation. Leveraging an annotated database for stem cell recognition, readily reconstitutable within an hour if needed, we'll design interfaces to facilitate AI training, stem cell state detection, and the precise delivery of growth factors. The project's specifics will be tailored in collaboration with the team, aligning with their expertise and preferences.
Dataset: Movies of developing stem cell aggregates
Programming Language & Toolbox: Python
For more information: https://drive.google.com/file/d/1mJt4n4hoWC8IfwiM37nEreGYzqiVNIgh/view?usp=sharing
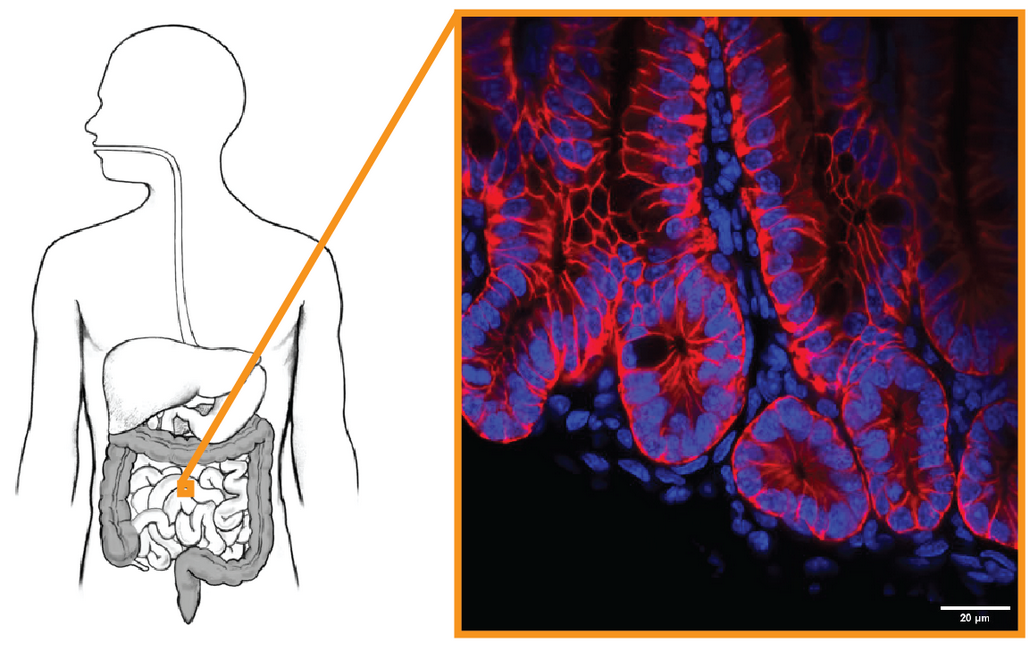
divCrypt: Automated segmentation in mouse intestinal crypt | Laura Ruiz, Prune Smolen
Keywords: Organoids, Computer Vision, Deep Learning
Digestion is supported by nutrient uptake through the very thin layer of the intestine. This layer, called intestinal epithelium monolayer, is constantly damaged by contacts with the heterogeneous substances circulating in our digestive system.
As such, the intestinal crypt continuously replenishes the epithelium with new cells to ensure its protection and continued function. Defects in the constant division process can be at the origin of the cryptogenesis problem as plural stratified epithelium leading to cancer. To study those defects, we grow mouse intestinal organoids composed of cryptes and systematically image and evaluate thousands of division events. However, this task is difficult to carry out due to high cell density and low signal-to-noise ratio. This problem challenges the robustness of the object segmentation, let alone actually uncovering the process of mitosis in a healthy intestinal epithelium.
This hackathon project will focus on the automated detection of cell division events in intestinal crypt images and investigate how cell rearrangements occur when a cell is added. This work will build upon manual annotation of the image dataset previously carried out in the laboratory. While participants will ample degree of freedom in the tool and milestone they favor, we suggest the following milestones that exploits and adapting previous work in machine learning for bioimaging:
- We will adapt recent work in the community (Karnat et al 2024) toward the detection of mitotic events in 2D+time nuclei images. We then combine this local approach with a nucleus segmentation tool (Weigert et al 2021) for a precise delineation of dividing nucleus.
- The membrane will then be detected on the membrane-dedicated channel using conventional contour detection toward the mapping of contacts points between diving nuclei and the closest membrane.
- Finally, the participant will be able to investigate the existence of patterns in the membrane/nuclei contact points in order to study the rearrangement strategy of newly appearing cells, as well as the computer-aided discovery of possible defects.
Useful Information
The event will take place from June 28 to June 30, 2024 at the Centre de séminaires Villages Clubs du Soleil, Belle de Mai, Marseille.
CENTURI Hackathon will welcome a maximum of 60 participants. Applicants should have some experience with coding.
Each participant should bring a laptop.
CENTURI will provide meals during the event (buffet) from June 28 to June 30.
CENTURI will cover accommodation and catering. CENTURI will not cover transportation nor additional costs.
Registration is free for all participants.
However, registered participants are expected to participate in the entire event (from June 28 early evening to June 30 early evening). After registering to the Hackathon, you will be contacted early May to select your projects.
For informal enquiries: info@centuri-livingsystems.org
Format
The CENTURI Hackathon will start on Friday, June 28, in the evening and will end on Sunday, June 30, at the end of the afternoon.
Deadline
Deadline for application: June 7, 2024
Venue
CENTURI Hackathon will take place in the Centre de séminaires Villages Clubs du Soleil, Belle de Mai, Marseille.

