CENTURI Hackathon
2023
CENTURI Hackathon
2023
Hacking Living systems
Joseph Aiguier CNRS research Institute
What is the CENTURI Hackhaton?
The CENTURI Hackathon is a fast-paced two-day event of coding, engineering and idea-sharing to drive innovations at the interface of Computer Science and Life Sciences. The event is aimed at curious and creative minds interested in improving their coding skills through collaborative projects. The event is open to students, postdocs, developers and researchers.
The goal of our Hackathon is to computationally unlock technological bottlenecks arising from the study of Living Systems. Our event is aimed at everyone with experience in coding and with a keen interest in problem solving in big data, computer vision or modeling. Experts in fields such as biology, mathematics and non-scientific developers are encouraged to apply!
Curious about the previous editions? Find out more about the 2022 Hackathon, and the 2023 Hackathon (Twitter Highlight 1 & Highlight 2) here!

What types of projects will participants be working on ?
Projects will strive to solve key technological bottlenecks in a variety of topics, such as Computer vision, Modeling, Data mining and Computer assisted microscopy.
Participants will work on big data projects for living systems, with projects from academic labs in CENTURI and elsewhere.
CENTURI Hackathon will welcome a maximum of 80 participants. Applicants should have some experience with coding.
Participants will work in groups of 6-8 people on various projects in a variety of topics:
- Data visualization and interactions
- Computer vision
- Data mining
- Robotics & Computer assisted microscopy
- Modelling
Projects are as follows:
FlyTrack: a flydome to understand muscle aging | Nuno Miguel Luis and Frank Schnorrer
Animals become progressively weaker as they grow older. The process behind the reduction in force production by the muscle is still poorly understood. Here, we use the fruit fly, Drosophila Melanogaster, and its extensive genetic toolkit to study changes in gene regulation and protein maintenance with age.
For this, we built a large arena called the Flydome, where flies can fly for long periods of time. To quantify their flight capacity with age we installed and synchronized 3 infrared cameras to record high frame rate videos of animals.
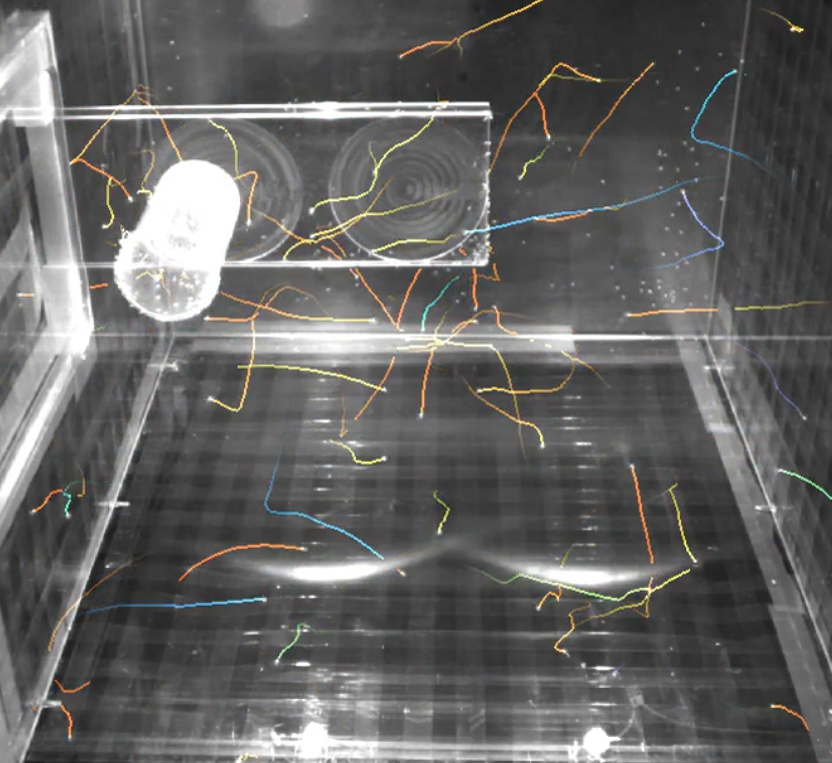
However, the large amount of insects, complex fly occlusion patterns and the sheer size of the raw data challenge reconstruction and understanding of the underlying aging process. To solve this computational challenge, we identified the following milestones along with associated benchmarks:
- Image processing and motion prediction to estimate 2D positions and displacements of flies in each of the 2D videos (see preliminary results in the illustration).
- Reconstructing 3D positions and displacements from multiple views acquired simultaneously.
- Long-term tracking, especially transitions from rest to flying, to quantify changes in flight capacity for each fly.
- Comparing fly behavior at different stages of their lifespans (eg. how and how much do young flies fly compared to old ones)
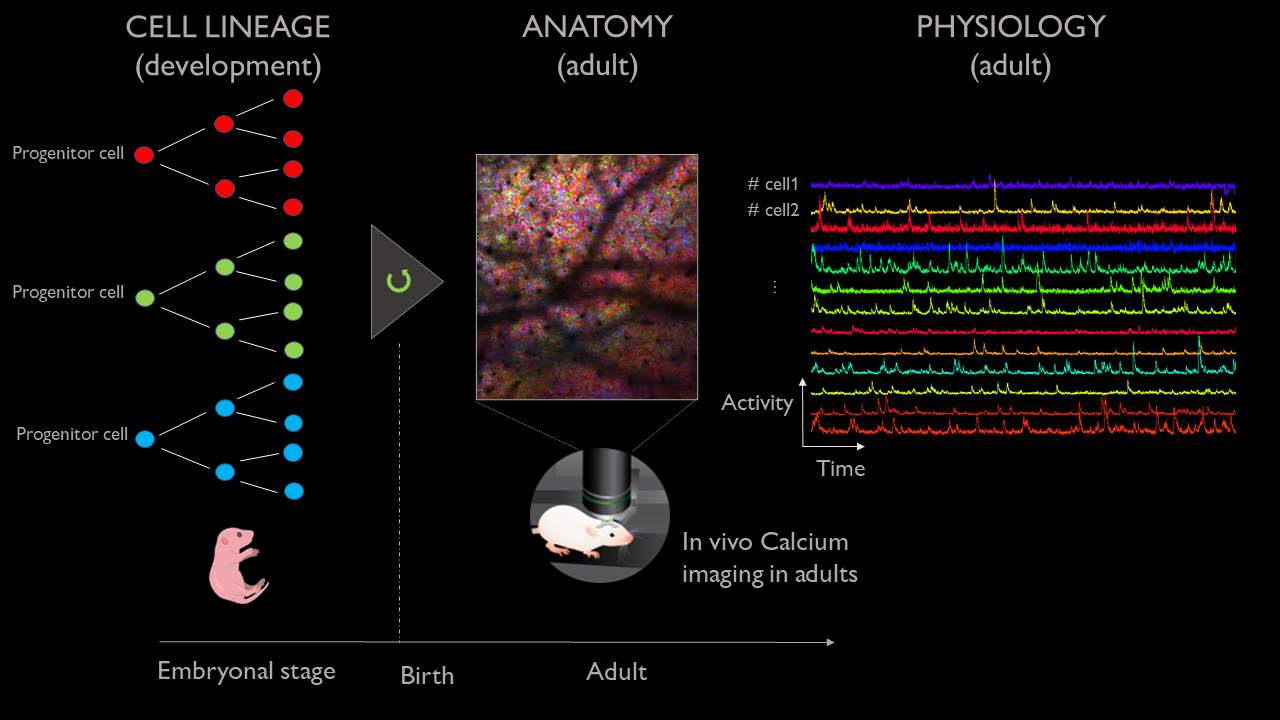
Taste the rainbow | Mirindra Ratsifandrihamanana and Jure Majnik
In the adult brain, neurons are active in groups called assemblies that orchestrate perception, action, and cognition. The genetic origin and functional homogeneity of cell assemblies and associated anomalies are still unknown. Investigating those processes requires the development of new tools that combined information from both neuronal activity as well as genetic lineage.
To do so, we developed an imaging technique that records the neuronal activity of living mice with cellular resolution using calcium-sensitive fluorescent dyes, combined with the labeling of each cell with a unique combination of colors that depends on their clonal origin (the ‘brainbow’ technique). Importantly, this technique can image the same mouse across multiple days. However, the resulting image sequences are complex, stochastic, and present changing geometry from one day to the next. As such, this project will focus on exploring the development of a few tools for the data-driven analysis of these multi-modal experiments. We have planned for the following tentative milestones representing increasing challenges.
- Associating brainbow signal and associated neuronal activity (calcium-recorded activity) at the cellular resolution using cell segmentation and co-localization across image modalities. This can be carried out using single-day imaging experiments.
- Interrogating the association between brainbow signal and neuronal activity. Is lineage (brainbow signal) predictive of neuronal activity (calcium-recorded activity) ? This can be carried out using single-day imaging experiments.
Matching cellular cluster from one day to the next through graph-matching techniques. Are all cells visible across all days? Is the association between brainbow signal and calcium signal stable across time?
BileFlow: Understanding the formation of hepatic tubules using organoids | Virgile Viasnoff
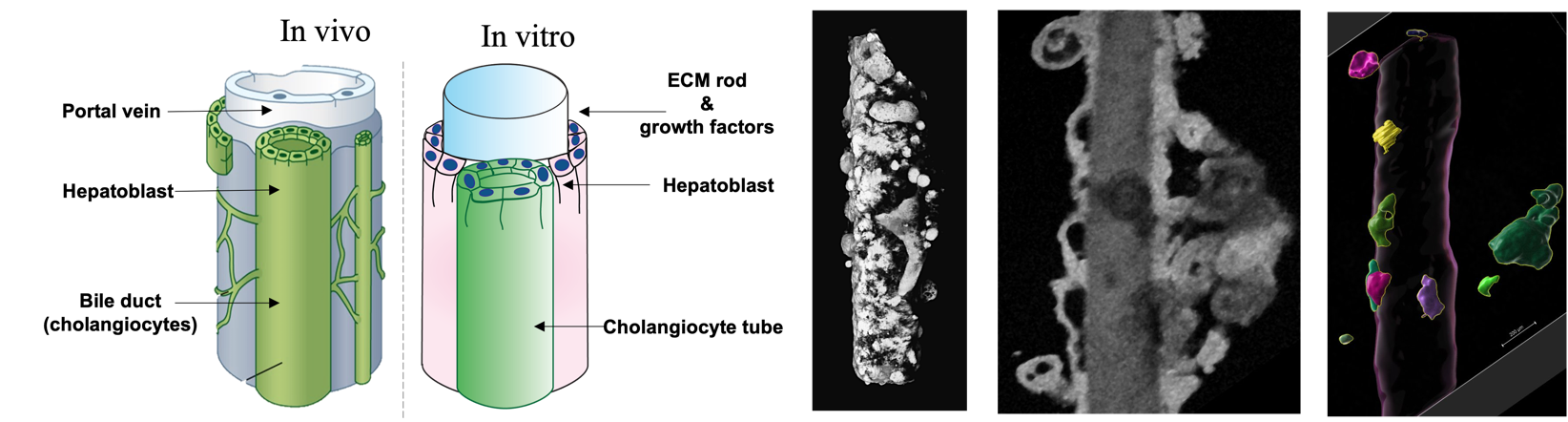
Bile is essential for digestion and detoxification. It is secreted by the liver and flows into the intestine in tubules with a hierarchical structure. The bile ducts, which collect the bile secreted by the hepatocytes, run along the portal veins. During embryonic development and liver regeneration, the ducts are formed by local differentiation of liver stem cells, which eventually form tubes running along the vein. While this developmental process is impaired in several diseases (such as alagile syndrome), it is also largely unknown.
To investigate this process, we used human induced pluripotent stem cells scaffolded by a bio-functionalized microrod to generate in vitro and in high throughput screening quantities of organoids that recapitulate the formation of the ducts. Using image data acquired from optical coherence tomography (OCT), our goal for this hackathon is to quantitatively analyze the complex transition undergone by spherical cellular aggregates into tubular ducts. In particular, a key question lies in the origin of bile ducts alignment along the main axis of the rods. We identified the following milestones to reach that goal:
1- To identify and segment the luminal cavities in 2D for each image plane. The challenge is to distinguish them from the rod and the background.
2- To perform a 3D reconstruction of the organoid duct system. This reconstruction will serve as a basis for live analysis of the timing of duct formation.
3- To classify the spherical luminal cavities (nascent) from the elongated ones (more mature) and to identify branching points.
4- To quantify the size and direction of growth of the tubular ducts.
Any success in those milestones would have an important impact on the understanding of synthetic organs. This impact could be broaden through the implementation of a batch analysis scheme compatible with the HCS format.
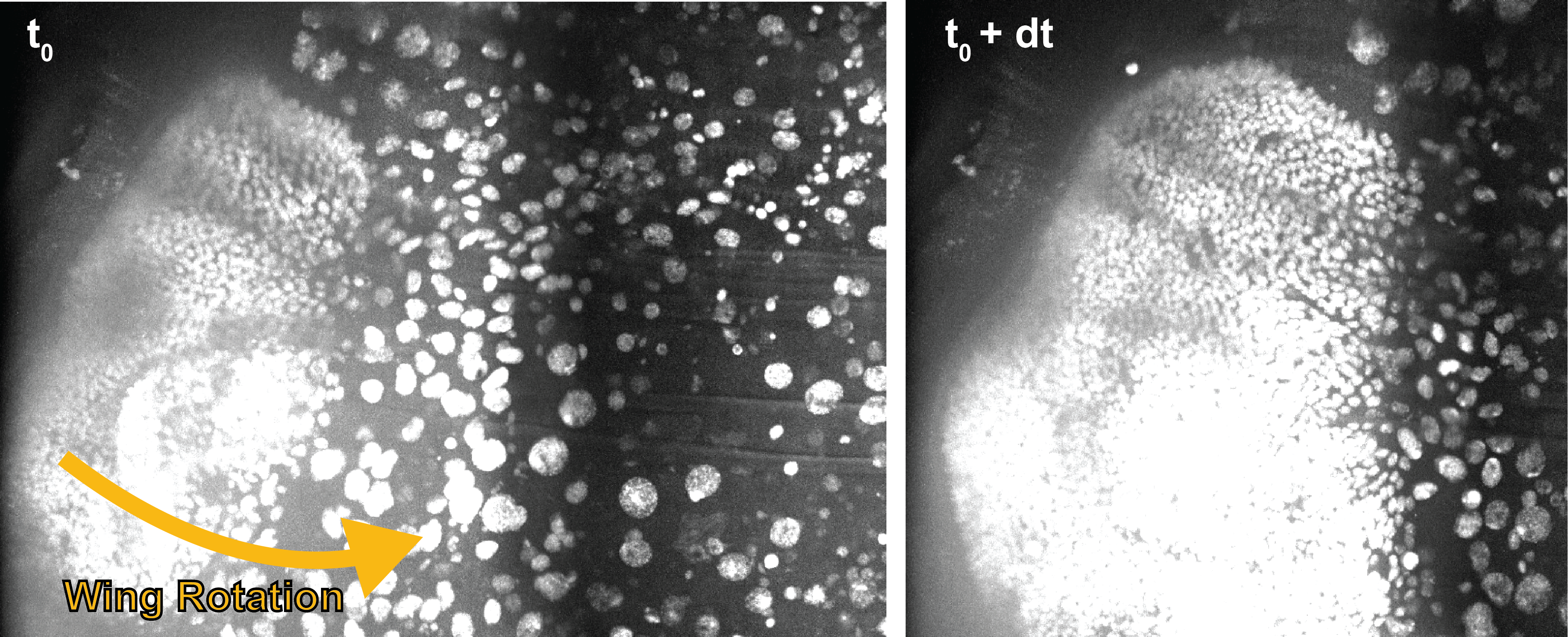
Ballerina: automated motion correction for minimally invasive imaging of developing tissues | Loic Legoff
Studying the microscopic processes that drive animal development is challenged by the large degree of freedom present during shapes formation.
We still lack a suitable technique to link the microscopic cellular processes that power morphogenesis to the global changes of shapes observed at the scale of the whole animal. This requires the observation at high spatio-temporal resolution of groups of cells which may move on large distances as the embryos change shape. Such a multi-scale problem is a challenge for the experimentalist: Using a low magnification imaging system to focus on large scale information, we may miss the essence of the cellular processes. Using a high magnification system to image cellular processes, we cannot follow the cells in time as they move out of the imaging field.
To tackle this problem, we will implement an adaptive microscope that uses a feedback to maintain a chosen group of cells at the center of the imaging volume as it moves on global scales. The feedback will also change the orientation of the sample in order to reach the best imaging conditions of the identified group of cells.
We mounted our sample on a stem attached to a rotating motor to follow the pupa as it moves. We are however facing two computational challenges in this task. First, one must detect the motion of the embryo for motor-driven correction. Second, we must take into account the delays and uncertainties in motor response for a steady view. As a first foray into this problem, our hackathon project will focus on the following milestones:
- Using conventional rigid transform technique, estimate the motion from one frame to the next.
- Test the approach through the difference between source and registered image.
- Is it robust to the heterogeneity of cell scale and motion types ?
- Simulating motor-driven rotation through isotrope sampling and projection.
- Simulate a motor response with the imprecision we measured in the lab in preliminary measurement.
- Build a proof of concept for motion estimation and correct the motors through a joint modeling of motion model and control uncertainty using conventional optimal control tools.
Heartoids: Exploring the formation of cardiac, vascular and intestinal domains in developing embryonic organoids | Sham Tlili
Embryonic organoids are in vitro multi-cellular aggregates that self-organize and reproduce the hallmarks of morphogenesis.
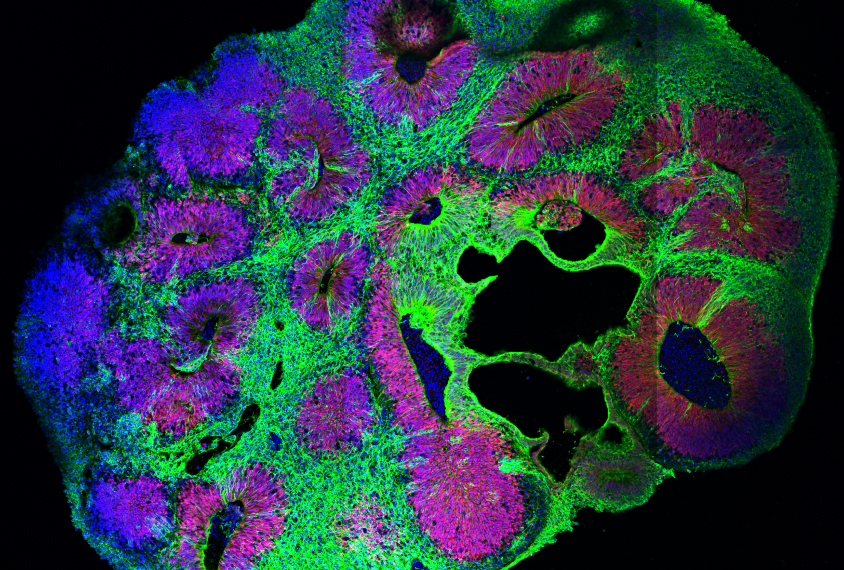
By setting stem cells in precise experimental conditions, our group and collaborators achieved the reproduction of cardiac functions in fully in vitro system, including the appearance of intestinal cavities, vascular tissues and cardiac domains with cardiac beatings. This new class of experiment enables the use of advanced microscopy and image analysis technics to understand the underlying biophysical processes shaping organs both during embryo development in vivo and in vitro.
To study this system, we use 2-photon volumetric imaging that reveals the cellular dynamic orchestrating organ formation in combination with late-stage staining to measure a larger number of genetic markers. The resulting 3D volumes present large cellular clusters with heterogeneous shapes and genetic expression. Visual observations readily yield new hypotheses such as a possible role of cell shapes coordination in local tissue function. However, the density and dimensionality challenge the study of such phenomena, let alone the discovery of less intuitive processes at play. As such, this project builds upon our preliminary work on deep-learning-aided cell detection, to link cell shape and genetic expression to drive the analysis of correlation between cellular morphology, tissular heterogeneity and their temporal hierarchy. The two-days hackathon project will be driven by the following questions:
- Interrogating correlation between cell morphology and tissue types that is suggested by visual inspection: Is cell density and tissue micro-structure predictive of tissue type (e.g. cardiac fibers versus cardiac progenitors)?
- Is this correlation also matched by a specific genetic landscape ?
- Can we combining cell morphology and genetic expression to determine new tissular and or functional clusters ?
- What comes first during heart formation: genes or shapes ?
Illustration courtesy of Institute of Molecular Biotechnology
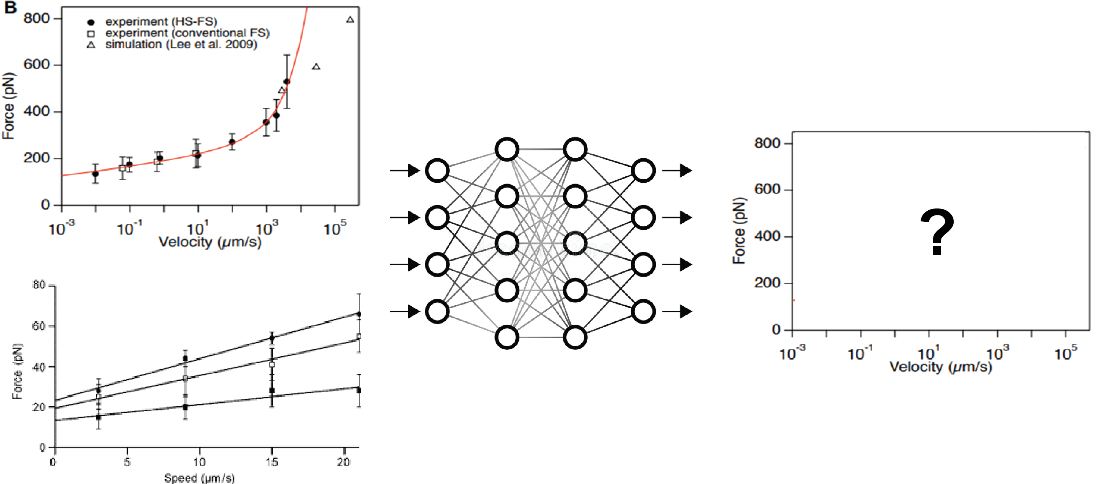
DeepLit Automated literature analysis applied to molecular stability prediction | Ismahene Mesbah and Felix Rico
The determination of protein structures is critical as it is directly linked to its function.
The emergence of AlphaFold 2.0 has recently demonstrated the prediction of protein structures from their sequences with the highest accuracy possible is possible. This was possible in part thanks to the large amount of structures available on the protein database (PDB), which feed the AlphaFold machine learning algorithm. Another crucial characteristic of proteins is its structural and mechanical stability, which, so far, cannot be predicted from its structure. For that a database of well-curated mechanical properties of proteins is needed. Such a database does not exist.
Available experimental and computational techniques permit to measure protein unfolding forces at different velocities (or loading rates). For example, Atomic Force Microscopy based Force Spectroscopy experiments permit to pull on single proteins and measure the forces required to unfold them. Computationally, Steered Molecular Dynamics (SMD) simulations is a computer simulation that mimics AFM pulling experiments and therefore provides an atomistic description of the unfolding process. While many papers have published in the field, no unified database is available. To address this issue, we need, first, to develop an automatic way to collect data that describes the mechanical stability of proteins. Concretely, the main objective of the project is to develop a Deep Learning algorithm that extracts data points from plots of unfolding forces versus loading rate and/or velocity curves, along the following milestones:
- Using annotations prepared before the hackathon, can we train a convolutional network to recover X/Y, and label from simple curve. The dataset comprises 30 annotated graphs of force versus loading rate or force versus velocity.
- Can we separate and identify different measurements within a plot from the legend? For example: experimental data (from AFM) and computational data (from SMD simulations)
- Can we link plot measuring the same quantities even with slightly different labels or legend?
Useful Information
The event will take place from June 14 to June 16, 2023 at the Joseph Aiguier CNRS research institute.
CENTURI Hackathon will welcome a maximum of 60 participants. Applicants should have some experience with coding.
Each participant should bring a laptop.
CENTURI will provide meals during the event (buffet) from June 14 to June 16.
CENTURI will not cover transportation, accomodation nor additional costs.
In case of financial difficulties for transportation and accomodation, you may contact us at info@centuri-livingsystems.org.
Registration is free for all participants.
However, registered participants are expected to participate in the entire event (from June 14 early evening to June 16 early evening).
For informal enquiries: info@centuri-livingsystems.org
Format
The CENTURI Hackathon will start on Wednesday, June 14, in the evening and will end on Friday, June 16, at the end of the afternoon.
Deadline
Deadline for application: June 9, 2023
Venue
CENTURI Summer School's will take place on the Joseph Aiguier CNRS research institute.

