Research
General overview and Research Groups
The Turing Centre aims to stimulate a highly interdisciplinary community, especially physicists, mathematicians and computer scientists who, together with biologists, provide the means to base experimental studies on solid quantitative grounds, to identify new questions and conceptualize new ways of understanding the complexity of living systems.
Life is based on dynamic systems whose organisational principles remain unclear due to their complexity. Such complexity has different origins. Living systems are the product of a long evolutionary history and are intrinsically dynamic as they develop, renew their components, adapt to their physiological environments and are trained. This is illustrated for instance in the development of embryos and organs, and in the function of the immune and nervous systems.
Understanding biological complexity is one of the most significant challenges for the future. It will impact both basic research and medicine, since diseases emerge from anomalies in the organisation and dynamics of living systems.
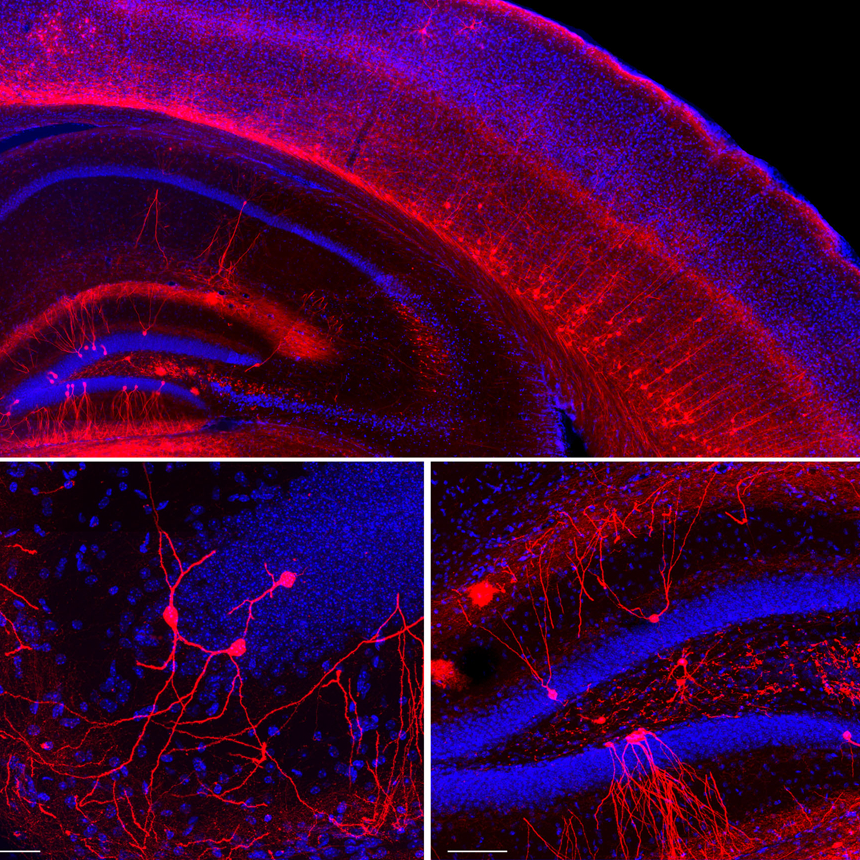
The Turing Centre addresses the nature of the information underlying biological processes, and the organisation and dynamics of biological structures. Five over-arching questions bring together biologists interested in development, neuroscience and immunology experts along with physicists, while computational biologists and mathematicians underlie the research effort:
How does genetic information impact cell fates?
How do signalling dynamics impact cell differentiation?
How is information encoded in collective cellular dynamics?
How do networks of cell interactions form and function collectively?
How does mechanics impact cell and tissue organisation and dynamics?
CENTURI recruits researchers and post-docs with backgrounds in computational biology, theoretical physics or biophysics, with a strong interest in working in a cross-disciplinary environment.
CENTURI fellows
Principal Investigators
Mathematical modelling of the ecology & evolution of host-associated microbiota
Brain mechanisms of flexible behavior
Understanding developmental reproducibility at the single cell scale
Self-organization and collective effects in living systems
Neural networks for the representation of 3D environments
Phillipe Roudot (Institut Fresnel)
Dynamic bioimage mining for live cell microscopy
Out-of-equilibrium Mechanics (OM)
Learning Meaningful Representations of Life
Florence Bansept (LCB) - Mathematical modelling of the ecology & evolution of host-associated microbiota
Lorenzo Fontolan (Inmed - CPT) - Brain mechanisms of flexible behavior
Léo Guignard (LIS) - Understanding developmental reproducibility at the single cell scale
Matthias Merkel (CPT) - Self-organization and collective effects in living systems
Hervé Rouault (CPT - INMED) - Neural networks for the representation of 3D environments
Pierre Ronceray (CPT) -Theoretical modeling and inference methods for the physics of soft biological matter
Phillipe Roudot (Institut Fresnel) - Dynamic bioimage mining for live cell microscopy
Jean-François Rupprecht (CPT) - Out-of-equilibrium Mechanics (OM)
Paul Villoutreix (LIS) - Marseille Machine Learning Team
Postdoctoral fellow
Mehdi Abbasi (CINaM, M2P2) -Understanding margination, from vessels to vascular networks
Ludovica Bachschmid Romano (Inmed) - Computational frameworks to link structure, dynamics and function in large-scale neural circuits
Maximilien Courgeon (IBDM, CPT) - Self-organising principles of Class IV neuron dendritic arborization in Drosophila
Paulina Garcia Gonzalez (IBDM) - EPIC-Mitochondria
Gesa Loof (LIS) - Early drivers of symmetry breaking in pre-implantation mouse embryo cell fate specification
Florian Labourel (LCB) - Metabolism and multi-stable equilibria in the gut microbiome
Sujit Kumar Nath (IBDM, CPT) - The effect of changing curvature on tissue flows in Drosophila embryo
Vitor Marquioni Monteiro (LCB) - Mathematical Modeling of Gut Microbiome
Ying Zhou (Inserm, TAGC) - Quantitive micromethods for rapid characterization of immune cells activity in infection and cancerous context
Sophie Carneiro Esteves (Fresnel)
Ludovica Bachschmid Romano (Inmed)
Computational frameworks to link structure, dynamics and function in large-scale neural circuits
Maximilien Courgeon (IBDM/CPT)
Self-organising principles of Class IV neuron dendritic arborization in Drosophila
Quantitive micromethods for rapid characterization of immune cells activity in infection and cancerous context
Sophie Carneiro Esteves (Fresnel)
PhD students
Ahmad Awada (LAI, I2M) - Reverse adhesive haptotaxis: from reductionist experimentation to mathematical and physical modeling
Alexandre Bonomo (CIML/LIS) - Multi-view machine learning analysis of intestinal immune response
Nadine Ben Boina (I2M, MMG) - Mathematical Modelling of rare diseases
Marie-Jose Chaaya (I2M, IBDM) Modeling tumor innervation: impact on immune cells and anti-cancer treatment
Jessica Chevallier (CIML, TAGC, LAI) - Spatial mapping of gene expression to decipher the thymic cellular
Arthur Coët MIO/CINaM) - Deciphering bacterial colonization of marine snow
Marie Dessard (CIML/LAI) - How cholesterol and phospholipids fine tune T cell early mechanosensitivity?
Jaime Fernandez MacGregor (TAGC) - Modeling human network perturbations by bacterial proteins
Andonis Gerardos (CINaM) - How to learn a stochastic partial differential equation from a video?
Alice Gros (IBDM, LIS) - Morphogenesis variability in embryonic organoids.
Mathias Ramm Haugland (IBDM/LIS) - MitoLearner: deep learning and explainable AI to understand the role of mitochondria genes in cancer cells
Andrei Karpov (AFMB/LAI) - Physical and structural characterization of the interplay between the cell membranes, actin filaments and Chikungunya protein nsP1
Martin Lardy (IRPHE,IBDM) - Computational Fluid Dynamics to infer embryonic tissue rheology
Emma Legait (IBDM) - Deciphering the self-organizing principles governing the cellular composition and growth of tumors
Marvin Leria (IBDM) - PCP plasticity in the basal metazoan Trichoplax adhaerens
Ismahene Mesbah (LAI, IBDM) - Decoding protein structural folds, sequence and structural motifs
Piyush Mishra (I2M, Institut Fresnel) - Weakly-supervised inferences of molecular dynamics for fluorescence imaging in physiological environments
Alexandre Ortega (LMA/LAI/IUSTI) - Development of acoustic techniques to study the biophysical properties of cell clusters
Harshit Pateria (IBDM/MMG) - Integrating transcription and lineage to define spatiotemporal trajectories of cardiac progenitor cells
Jessica Pereira Silva (INMED/I2M) - Behavioural grammar and synaptic rules of the social brain
Marc-Eric Perrin (IBDM, CPT) - Models of neuronal growth during embryogenesis as self-interacting dynamical trees
Devam Purohit (CINaM, I2M) - The muscle spring: quantifying and manipulating the molecular elasticity of muscle in vitro and in vivo
Florian Saby (IRPHE/LCB) - Understanding Decision-Making in Myxococcus xanthus
Mirindra Ratsifandrihamanana (INMED, CPT) - Circuits supporting dynamics transitions in the developing hippocampus
Hilde Langengen Teigen (INMED, INT) Neuromodulatory mechanisms of predictive processing in the mouse visual cortex
Rémy Torro (CINaM, LAI) - Mapping and classifying the topography of adhesive cell interface with high precision
Paul Uteza (CPT, INMED) - Neural networks for the representation of 3D environments
João Pedro Valeriano Miranda (CINaM) - Learning hidden structures in the stochastic dynamics of living systems
Jules Vanaret (I2M, IBDM) - OrganoDyn - Organoid dynamic reconstruction across scales
Matthew Jones (LCB)
Claire Plateaux (MMG/I2M)
Corentin Gourc (LMA/IBDM)
Tom Orjollet-Lacomme (Inmed/IRPHE)
Nicole Kolodziej (Indmed)
Louis Ritchi (Inmed)
Sujal Kataria (DyNaMo/IBDM)
Jad Sleiman (CIML/LAI)
Reverse adhesive haptotaxis: from reductionist experimentation to mathematical and physical modeling
Multi-view machine learning analysis of intestinal immune response
Mathematical Modelling of rare diseases
Modeling tumor innervation: impact on immune cells and anti-cancer treatment
Jessica Chevallier (CIML/TAGC/LAI)
Spatial mapping of gene expression to decipher the thymic cellular
Deciphering bacterial colonization of marine snow
How cholesterol and phospholipids fine tune T cell early mechanosensitivity?
How to learn a stochastic partial differential equation from a video?
Mathias Ramm Haugland (IBDM/LIS)
MitoLearner: deep learning and explainable AI to understand the role of mitochondria genes in cancer cells
Physical and structural characterization of the interplay between the cell membranes, actin filaments and Chikungunya protein nsP1
Deciphering the self-organizing principles governing the cellular composition and growth of tumors
Piyush Mishra (I2M/Institut Fresnel)
Weakly-supervised inferences of molecular dynamics for fluorescence imaging in physiological environments
Alexandre Ortega (LMA/LAI/IUSTI)
Development of acoustic techniques to study the biophysical properties of cell clusters
Integrating transcription and lineage to define spatiotemporal trajectories of cardiac progenitor cells
Models of neuronal growth during embryogenesis as self-interacting dynamical trees
The muscle spring: quantifying and manipulating the molecular elasticity of muscle in vitro and in vivo
Mirindra Ratsifandri-Hamanana (INMED/INT)
Circuits supporting dynamics transitions in the developing hippocampus
Hilde Langengen Teigen (INMED/ INT)
Neuromodulatory mechanisms of predictive processing in the mouse visual cortex
João Pedro Valeriano Miranda (CINaM)
Learning hidden structures in the stochastic dynamics of living systems
Matthew Jones (LCB)
Baya Djadoun (IBDM)
Claire Plateaux (MMG/I2M)
Corentin Gourc (LMA/IBDM)
Tom Orjollet-Lacomme (Inmed/IRPHE)
Nicole Kolodziej (Inmed)
Louis Ritchi (Indmed)
Sujal Kataria (DyNaMo/IBDM)
Jad Sleiman (CIML/LAI)
Yan Plötze (IBDM/CPT)
Engineers
Lea Chabot - Bioinformatics Engineer
Audrey Comte - Software Development Engineer
Fabrice Dessolis - Mechatronics Engineer
Thang Duong Quoc Le - Image Data Processing & Analysis
Mai Han Hoang - Image Data Processing & Analysis
Mathias Lechelon - Mechatronics
Stefania Sarno - Data Neuroscientist
Thomas Vannier - Bioinformatics Data Analyst
Alumni
Since 2017, talented and innovative scientists have helped us shape CENTURI and collaborate on federating a diverse community and to ensure its commitment to an interdiciplinary approach of Science and Research.
Photography by ©Criscuolo