PHD2024-24
Neural Tissue patterning: how tissue mechanics feedback on genetically encoded programs
Host laboratory and collaborators
Mehdi Saadaoui, IBDM mehdi.saadaoui@univ-amu.fr
Raphaël Clément, IBDM raphael.CLEMENT@univ-amu.fr
Abstract
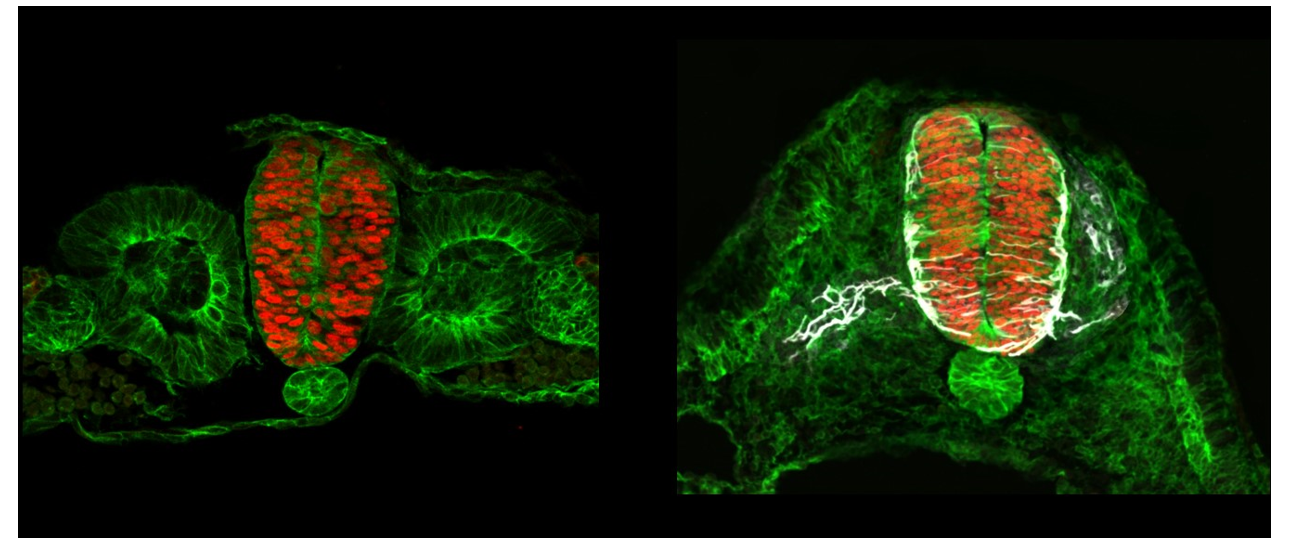
In the central nervous system, tissue patterning is well documented at the molecular level but remains largely unexplored from a biophysical perspective. We particularly lack information about the role played by cell and tissue mechanics. For example, Dorso-ventral patterning of the neural tube relies on the integration of two opposite morphogen gradients, i.e Shh signaling coming from the notochord/floor plate and BMP signaling coming from the roof plate. In the lab, we recently observed supracellular actomyosin cables restricted to specific subdomains along the D-V axis. Our aim is to investigate whether the different progenitor populations arising from the D-V patterning present different mechanical properties and if cell and tissue mechanics participate in the creation and/or maintenance of sharp boundary between these different domains by a spatial mechano-transcriptomic approach. We plan to combine a detailed deep-learning-based morphometric and mechanical analysis of the tissue with genetic patterning data, in order to extract the mechanical parameters that vary according to genetic patterning.
Keywords
Neuroepithelium, tissue patterning, cell and tissue mechanics, multi-scale quantitative analysis
Objectives
Our objective is to:
- Obtain a morphometric characterization of the developing neuroepithelia
- Analyze the correlation between tissue mechanics and genetic information
- Modulate the candidate parameters identified in aim 1 in order to assess how they impact the correlation inferred in the aim 2
Proposed approach (experimental / theoretical / computational)
The candidate will carry out experimental and computational work.
He/She will develop an analysis pipeline to extract quantitative morphometric and mechanical information using datasets from fixed and live-imaged neuroepithelial tissue (some are already available) in order to obtain an integrative representation of the developing neuroepithelium. Morphometry will be assessed using automated image analysis, while mechanical information will be assessed using Bayesian force inference. The mechanical and morphological cell-scale information will be combined and analyzed using K-means clustering in order to define territories based on these characteristics. To link this map to genetic information, the candidate will use single-cell transcriptomic data from the spinal cord to correlate differentially expressed genes to regions of the neuroepithelium with different mechanical/morphological properties.
He/She will then use genetic, chemical or mechanical tools available in the lab in order to perturb cell/tissue mechanics, and use the pipeline to analyze if some perturbations affect tissue boundary sharpness.
Interdisciplinarity
This project is at the interface between developmental biology, neuroscience, and tissue mechanics. The objective is to bridge the gap between tissue mechanics and cell fate specification in the context of the vertebrate central nervous system morphogenesis.
To that end, we plan to use state-of-the-art image analysis techniques, Bayesian inference to infer cellular forces, K-means clustering, and single cell transcriptomics. We anticipate that this highly interdisciplinary combination of approaches will provide unparalleled insights into the mechanisms that relate the physical state of the tissue to its genetic patterning.
The candidate will be co-supervised by a biologist (Mehdi Saadaoui) and a physicist (Raphaël Clément), both localized in different teams of the Institute of Developmental Biology of Marseille (IBDM). He/she will share the time between the two labs.
In the biology lab, the candidate will carry out experiments and data analysis.
In the modeling lab, he/she will develop the pipeline to combine morphometric and mechanical data to perform K-means clustering, and eventually compare it to genetic data obtained in his/her experiments. If time allows, and if the data justifies it, the candidate may participate in the development of a dynamic model relating tissue patterning to its physical state.
Expected profile
The candidate should have a Master (or international equivalent) in experimental physics, biophysics or biology with computer programming skills (Python/Matlab). Although basic notions of developmental biology will be an asset, they are not essential. The candidate must be prepared to divide his time between wet and dry lab and to work with avian embryos..
2 to 5 references related to the project
- Regulation of size and scale in vertebrate spinal cord development. Kuzmicz-Kowalska K, Kicheva A. Wiley Interdiscip Rev Dev Biol. 2021 May;10(3):e383. doi: 10.1002/wdev.383. Epub 2020 May 11. PMID: 32391980
- Neuronal differentiation influences progenitor arrangement in the vertebrate neuroepithelium. Pilar Guerrero, Ruben Perez-Carrasco, Marcin Zagorski, David Page, Anna Kicheva, James Briscoe, Karen M Page. Development 2019 Dec 4;146(23): dev176297.
- A computational pipeline for spatial mechano-transcriptomics. Adrien Hallou, Ruiyang He, Benjamin D. Simons, Bianca Dumitrascu. BioRxiv doi: https://doi.org/10.1101/2023.08.03.551894
Two main publications from each PI over the last 5 years
- Serotonin signaling regulates actomyosin contractility during morphogenesis in evolutionarily divergent lineages. Karki S, Saadaoui M, Dunsing V, Kerridge S, Da Silva E, Philippe JM, Maurange C, Lecuit T. Nat Commun. 2023 Sep 8;14(1):5547. doi: 10.1038/s41467-023-41178-w.
- A tensile ring drives tissue flows to shape the gastrulating amniote embryo. Saadaoui M, Rocancourt D, Roussel J, Corson F, Gros J. Science. 2020 Jan 24;367(6476):453-458. doi: 10.1126/science.aaw1965.
- RhoA- and Cdc42-induced antagonistic forces underlie symmetry breaking and spindle rotation in mouse oocytes. Dehapiot B, Clément R, Bourdais A, Carrière V, Huet S, Halet G. PLoS Biol. 2021 Sep 7;19(9):e3001376. doi:10.1371/journal.pbio.3001376. eCollection 2021 Sep.
- Experimental validation of force inference in epithelia from cell to tissue scale. Kong W, Loison O, Chavadimane Shivakumar P, Chan EH, Saadaoui M, Collinet C, Lenne PF, Clément R. Sci Rep. 2019 Oct 10;9(1):14647. doi: 10.1038/s41598-019-50690-3.
Project's illustrating image