PHD2024-20
Probing Biomechanical Interactions in 3D Intestinal Organoid Development
Host laboratory and collaborators
Loïc Le Goff, Institut Fresnel, loic.le-goff@univ-amu.fr
Felix Rico, DyNaMo, felix.rico@inserm.fr
Delphine Delacour, IBDM, delphine.DELACOUR@univ-amu.fr
Abstract
Organs self-organize into structured 3D shapes that serve their function. Addressing the complex biomechanics of tissue morphogenesis requires the development of new experimental approaches to assess cell-cell and cellmatrix interactions in complex 3D assemblies.
In this project, we will develop an experimental paradigm to link the 3D shape of intestinal organoids to cell-cell
and cell-matrix chemical and mechanical signaling.
We will investigate the biomechanical interactions at play by combining 2D and 3D intestinal organoids that are amenable to high-resolution (HR) imaging and mechanical characterization, and HR-traction force microscopy using super-resolved fluorescence imaging and viscoelastic mapping with atomic force microscopy. We will use novel 3D optical imaging to decipher the organoid cytoskeleton arrangements, map tissue tensions through laser dissection, and understand the ECM remodeling during intestinal growth. This multi-modal analysis will relate the global shape with active forces from the cytoskeleton, traction- and remodeling-forces exerted on the ECM.
Keywords
Intestinal organoids; smart-microscopy; super-resolution microscopy; atomic-force microscopy; traction force
microscopy; morphogenesis;
Objectives
The main objective is to correlate organoid 3D shape with cytoskeletal architecture and traction forces on ECM. Using 3D imaging and mechanical assessments, we will explore niche and stem cell roles at the organoid crypt, studying their interactions with ECM. This analysis will intertwine with HR-mechanical assessment on 2D organoid-derived monolayers, which share many features of the organoid. This 2D system allows for AFM, HRtraction force microscopy, and super-resolution fluorescence microscopy, facilitating an in depth mechanical exploration of crypt cell types and their ECM interactions.
Proposed approach (experimental / theoretical / computational)
We will use state-of-the-art approaches in mechano-biology and imaging developed by our teams:
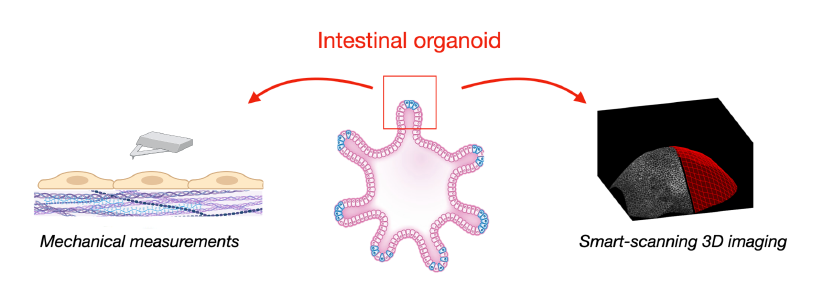
Intestinal organoids are popular in vitro models that grow in hydrogel-based 2D or 3D substrates [1,2,5,6]. They emulate 'mini-organs' that faithfully replicate numerous physiological and architectural features of real intestines and are amenable to live imaging and mechanical measurements.
3D imaging will be performed with our recently developed smart-scanning microscope [3,7,8], which focuses the imaging process on the structure of interest of the organoid, for example its surface. The benefit is high-resolution imaging with an increased speed and reduced phototoxicity.
We will infer traction forces from strain measurements of the embedding ECM (traction force microscopy). This will be complemented with second harmonic generation (SHG) microscopy to assess reorganization of the ECM. On the 2D system, we will improve traction force resolution through super-resolved imaging developed in the lab [8].
AFM of 2D organoids and the underlying matrix will provide quantitative information of the mechanical
properties of the system which will be used to refine the traction forces derived from the deformation
maps in 3D. Moreover, it will allow us to determine the viscoelastic state of the various cell types.
Interdisciplinarity
This project builds on the complementary expertises of three teams.
- The MOSAIC Team at Institut Fresnel (L. Le Goff) specializes in 3D imaging of biological tissues. Their recent development in smart microscopy and super-resolved microscopy will be an asset to image curved organoids, track fiduciary markers in the ECM for traction force, and assess the ECM structure around the
organoids. - The team E2M at IBDM (D. Delacour), focuses on understanding epithelial morphogenesis, homeostasis, and intestinal pathologies, with expertise in cell dynamics, particularly in 2D and 3D mammalian model systems. The project builds on their expertise in 2D and 3D organoid cultures.
- The AFM group at DyNaMo lab (F. Rico) specializes in the mechanics of biomolecules, cells and tissues. Their recent development in viscoelastic mapping using AFM will be essential for a thorough characterization of the mechanics of organoids and the surrounding ECM.
The student will thus thrive in a highly interdisciplinary environment, building expertise in fields as diverse as optics, biomechanics and developmental biology.
Expected profile
We seek a student in biophysics with a will to delve in the implementation of novel experimental tools for tissue mechanics. Programming skills will be an important asset.
Is this project the continuation of an existing project or an entirely new one? In the case of an existing project, please explain the links between the two projects
This is a new project that will build on the strong expertise of the 3 labs: Optical microscopy at Institut Fresnel, Force microscopy at DyNaMo and organoid development at IBDM.
2 to 5 references related to the project
- Xi, Saleh, Yamada, Mercier, Dang, Janel, Soleilhac, Wu, Tomba, Romagnolo, Lafont, Mège, Chen and Delacour. Modulation of designer biomimetic matrices for optimized intestinal epithelial cultures. Biomaterials (2022), 282:121380. doi: 10.1016/j.biomaterials.2022.121380
- Digesting the mechanobiology of the intestinal epithelium. Perez-Gonzalez et al. (2022). Current Opinion in Genetics & Development dot: 10.1016/j.gde.2021.10.005
- Meng, Nuzdhin, Sison, Galland, LeGoff (2023). Adaptive scans allow 3D-targeted laser-dissection to probe the mechanics of cell sheets. EPJ Plus, https://rdcu.be/dj9Ms DOI : 10.1140/epjp/s13360-023-04378-3
- Ackermann, Qu, LeGoff*, Ben Amar*. (2022) Modeling the mechanics of growing epithelia with a bilayer plate theory. EPJ Plus 137:8. DOI:10.1140/epjp/s13360-021-02205-1
Two main publications from each PI over the last 5 years
Delacour:
- Gaston, De Beco, Doss, …, Lim, Ladoux and Delacour (2021). EpCAM promotes endosomal modulation of the cortical RhoA zone for epithelial organization. Nat Commun 12, 2226. doi: 10.1038/s41467-021-22482-9
- Saleh, Fardin, Barai, …, Minc* and Delacour* (2023). Length limitation of astral microtubules orients cell divisions in murine intestinal crypts. Dev Cell 58, 1519-1533 e1516. doi: 10.1016/j.devcel.2023.06.004
LeGoff:
- Mazzella, Mangeat, Giroussens, Rogez, Li, Creff, Saadaoui, Martins, Labouesse, Idier, Galland, Allain, Sentenac, LeGoff (2023). Extended-depth of field random illumination microscopy, EDF-RIM, provides superresolved projective imaging. BioRxiv DOI: 10.1101/2023.10.30.564754.
- Abouakil, Meng, Burcklen, Rigneault, Galland, LeGoff (2021). An adaptive microscope for the imaging of biological surfaces. Light: science and applications 10, 210 (2021). DOI: 10.1038/s41377-021-00649-9
Rico:
- López-Alonso, Eroles, Janel, Berardi, Pellequer, Dupres, Lafont, and Rico. 2023. PyFMLab: Open-source software for atomic force microscopy microrheology data analysis. Open Research Europe. 3. doi:10.12688/openreseurope.16550.1
- Eroles, Lopez-Alonso, Ortega, Boudier, Gharzeddine, Lafont, Franz, Millet, Valotteau, and Rico. 2023. Coupled mechanical mapping and interference contrast microscopy reveal viscoelastic and adhesion hallmarks of monocyte differentiation into macrophages. Nanoscale. 15:12255–12269. doi:10.1039/D3NR00757J
Project's illustrating image