PHD2024-12
Imprint of mechanical forces on antibody affinity maturation in B cell immune responses
Host laboratory and collaborators
Philippe Robert, LAI philippe.robert@inserm.fr
Pierre Milpied, CIML milpied@ciml.univ-mrs.fr
Abstract
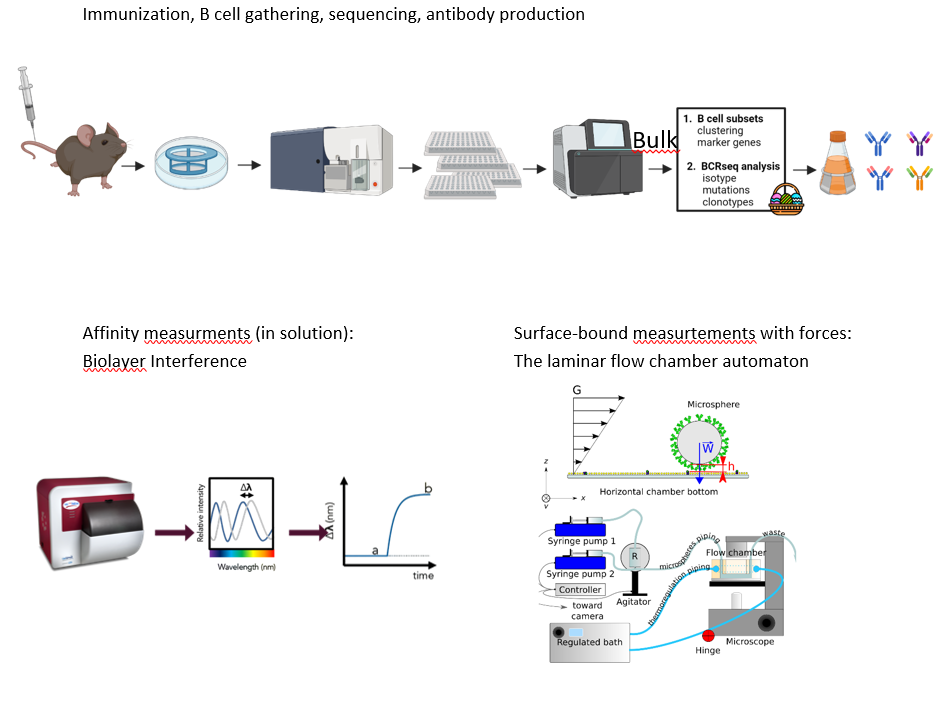
In the course of an immune response, the progeny of a single antigen-specific B lymphocyte generates highly potent antibodies through iterative cycles of (i) de novo mutations in the antibody coding genes and (ii) of selection of B cells expressing mutated antibodies through competitive antigen binding. Recent research challenges the assumption of clear affinity increases during this process. Our recent findings indicate that, in an ovalbumin immunization model, mechanical force resistance plays a crucial role in driving antibody selection, even in the absence of observable affinity gains. We aim to extend this result by studying responses to diverse antigens exhibiting structural variations, such as multivalency. We have enhanced our experimental setup to incorporate a broader range of physical parameters, for faster and more comprehensive results. We will quantify the binding properties of antibody lineages both in solution and under force, while also assessing the functional impact of mutations by reverting them. We intend to create a unique dataset that serves as a model to elucidate the relationship between antibody sequences and their biophysical binding characteristics.
Keywords
Antibody, affinity maturation, B lymphocyte, force, single bond
Objectives
Firstly (objective 1), we aim to extend the concept of force-driven antibody maturation to novel antigens. To achieve this, we will characterize antibody-antigen interactions at the single-molecule level in a two-dimensional setting, applying diverse forces to antibodies derived from B cells at various stages of affinity maturation in vivo. Utilizing innovative features of our main method, (objective 2), we intend to compile a comprehensive single-molecule multiparametric dataset of antibody-antigen binding to establish a direct link between the sequence, structure, and function of these molecular bonds.
Proposed approach (experimental / theoretical / computational)
In mice intentionally vaccinated or infected with several different antigenic models, we will sort antigen-specific B cells undergoing affinity maturation for parallel analysis of transcriptome and antibody genes sequences at single-cell resolution. We will produce recombinant antibodies derived from B cells of common ancestry at various stages of affinity maturation, then assess effect of mutations by reverting them.
Our laminar flow chamber system measures the current largest datasets of single bond life-times under forces; it was recently developed to measure uniquely new parameters at single molecular level: association kinetics, binding energies and bond ageing. Automatization of sample preparation and data analysis will allow to gather larger datasets.
Relationships between maturation stage, antibody structural features and binding properties for each lineage of antibodies will ultimately be formalized thanks to an ongoing collaboration with J-F Rupprecht (CPT).
Interdisciplinarity
The project unites two complementary CenTuri teams. Philippe Robert's team at LAI specializes in the development of laminar flow chambers for precisely measuring the binding properties of TCR to pMHC and antibodies to antigens. Leveraging highly automated methods, the team measures extensive collections of ligand-receptor interactions. LAI's work includes the formalization of binding interactions between proteins, providing a theoretical foundation to describe the physical properties of these molecular bonds.
Pierre Milpied's team at CIML focuses on the biology of B cell responses, employing cutting-edge single-cell genomics tools. Their adeptness lies in selecting maturing B cells, producing their antibodies, and conducting comprehensive genetic and structural characterizations. The team brings valuable experience in the quantitative analysis of B cell differentiation dynamics derived from single-cell gene expression data.
Together, these teams blend expertise in state-of-the-art biophysical measurements with in-depth knowledge of single-cell genomics, poised to advance the understanding of ligand-receptor interactions and B cell responses.
Expected profile
Tasks will consist in producing antibodies, preparing reactive surfaces for flow chamber experiments, performing flow chamber experiments, extracting and compiling binding data through dedicated software and discussing results.
The candidate must either be a cellular biologist or immunologist with a strong interest in biophysics, or a physicist with a strong interest in biology and immunology.
Is this project the continuation of an existing project or an entirely new one? In the case of an existing project, please explain the links between the two projects
This project is the continuation of a 2020 project by Pierre Milpied and Philippe Robert that succeeded in showing the role of resistance to force over affinity during the selection process of antibody maturation after ovalbumin immunization (manuscript in preparation). The present aim at building on methodological upgrades to extend the biological result to other immunizations and gather more quantitative parameters to understand the biophysics of matured antibodies binding. The project will be funded partly by ANR PRC grant “GC selection” (2024-2027, PI P.Milpied, partner P.Robert).
2 to 5 references related to the project
- Germinal centers output clonally diverse plasma cell populations expressing high- and low-affinity antibodies. Adrien Sprumont, Ana Rodrigues, Simon J. McGowan, Colin Bannard, Oliver Bannard Cell . 2023 Dec 7;186(25):5486-5499.e13. doi: 10.1016/j.cell.2023.10.022
- Immune cells use active tugging forces to distinguish affinity and accelerate evolution. Hongda Jiang and Shenshen Wang March 10, 2023 Proc Natl Acad Sci U S A. 2023 Mar 14;120(11):e2213067120.doi: 10.1073/pnas.2213067120
2 main publications from each PI over the last 5 years
- Limozin L, Bridge M, Bongrand P, Dushek O, van der Merwe PA, Robert P TCR-pMHC kinetics under force in a cell-free system show no intrinsic catch bond, but a minimal encounter duration before binding. Proc Natl Acad Sci U S A. 2019 Aug 20;116(34):16943-16948.doi: 10.1073/pnas.1902141116.
- Johannes Pettmann, Lama Awada, Bartosz Różycki , Anna Huhn, Sara Faour, Mikhail Kutuzov, Laurent Limozin, Thomas R. Weikl, P. Anton van der Merwe*, Philippe Robert *†(† :co-dernier auteur) and Omer Dushek *† Mechanical forces impair antigen discrimination by reducing differences in T cell receptor off‐rates. EMBO Journal 2023, doi : 10.15252/embj.2022111841 PMID : 36484367.
- Gregoire C, Spinelli S, Villazala-Merino S, Gil L, Holgado MP, Moussa M, Dong C, Zarubica A, Fallet M, Navarro JM, Malissen B, Milpied P*, Gaya M*. Viral infection engenders bona fide and bystander subsets of lung-resident memory B cells through a permissive mechanism. Immunity. 2022. doi: 10.1016/j.immuni.2022.06.002. *co-corresponding author
- Attaf N, Cervera-Marzal I, Dong C, Gil L, Renand A, Spinelli L, Milpied P. FB5P-seq: FACS-based 5-prime end single-cell RNA-seq for integrative analysis of transcriptome and antigen receptor repertoire in B and T cells. Front Immunol. 2020 Mar 3;11:216. PMID: 32194545.
Project's illustrating image