PHD2024-06
Neuropeptide control of epithelial dynamics in the simple animal Trichoplax adhaerens
Host laboratory and collaborators
Andrea PASINI, IBDM andrea.pasini@univ-amu.fr
Raphaël CLEMENT, IBDM raphael.clement@univ-amu.fr
Philippe ROUDOT, Institut Fresnel philippe.roudot@univ-amu.fr
Abstract
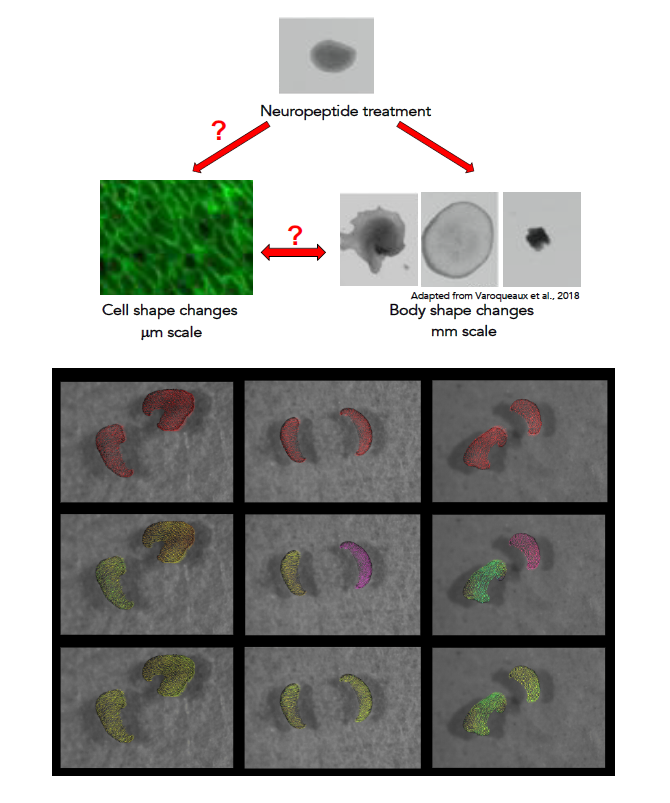
Trichoplax adhaerens is a small marine animal with a flat body (about 2mm in diameter and 25μm thick) composed of a few thousand cells organised in two epithelial sheets (1). One striking feature of Trichoplax is its morphological plasticity, single individuals being able to dramatically change their body shape in a few minutes (1-3). Despite the absence of a defined nervous system, several neuropeptides belonging to different families (Insulin-like, Endomorphin-like, YYamide, RWamide, SIFGamide, etc) are produced by scattered secretory cells and modulate the animal shape and behaviour (4, 5). The project aims at exploring how neuropeptides affect the shape changes and collective movements of Trichoplax epithelial cells, eliciting distinct, rapid and reversible effects such as shrinking, flattening and increase or arrest of the intra-epithelial cell movements. We will exploit a panel of experimental, computational and modelling approaches to study how different selected neuropeptides affect the shape and collective behaviour of Trichoplax epithelial cells.
Keywords
Epithelial dynamics; Cell shapes changes; Collective behaviours; Neuropeptides; Modelling data analysis; Multiple object tracking
Objectives
The project aims at using a combination of experimental, computing and modelling approaches to explore how treatment with different neuropeptides affects epithelial cell shape and collective behaviour in the morphologically highly plastic, early diverging metazoan Trichoplax adhaerens. This study will shed light on the evolutionary history of neural signalling in animals, and on how complex, coordinated collective cell behaviours can be maintained in the absence of a connected neural network.
Proposed approach (experimental / theoretical / computational)
Animals will be treated with variable concentrations of neuropeptides known to induce clear whole-body effects: LF-amide (flattening); PWN-amide (shrinking); FFNP-amide (increased epithelial movements) and LFNE-amide (arrest of epithelial movements) (4). The outcome of the different treatments on animal and cell motility will be assessed with a software for unsupervised tracking developed by P. Roudot and A. Pasini. Treated animals will also be fixed, subjected to IF with an antibody against MAGUK (Membrane-Associated GUanilate-Kinase) or with fluorescent phalloidins to stain the epithelial cells outlines and actin cytoskeleton (1), then imaged in confocal microscopy and segmented to measure different cell parameters and shape descriptors. The underlying tissular mechanical states will be assessed with a Bayesian force inference approach. Also, live animals stained with Cell Mask Orange (cell outline) and SiR-actin (actin cytoskeleton) will be filmed by Spinning Disc Roper confocal videomicroscopy. This interdisciplinary approach will inform on the cellular bases of neuropeptide-induced wholebody shape changes.
Interdisciplinarity
The project highly interdisciplinary as it combines experimental biology, advanced tracking approaches and mathematical modelling. This combination has never been applied to Trichoplax and promises to shed light on the individual cell- and collective behaviours underlying the whole-body shape changes induced by neuropeptides in this basally-diverging animal. The supervisors of the project have complementary background to cover project mentoring and significant experience in interdisciplinary research : developmental and cell biology of aquatic organisms for A. Pasini, in silico modelling applied to biological problems for R. Clément and computer vision methods for analysis of biodynamics for P. Roudot. Recent collaboration among the three supervisors have proven very fertile. A.Pasini /R. Clément: co-supervision of C. Rulquin, post-doc (Chuyen et al. Dev. Cell 2021); ongoing co-supervision of M. Leria, PhD student. A. Pasini/P.Roudot: co-supervision of L. Neschen, M2 student.
Expected profile
Applicants should have a background in either biology or physics/biophysics, with a strong interest for computational biology. The project is suitable for biologists with a strong interest in quantitative biology, or for physicists with a strong motivation to carry out biological experiments. A passion for evolutionary studies of marine organisms and previous knowledge of neuropeptide signalling would be an advantage. Ability to communicate in English is mandatory.
Is this project the continuation of an existing project or an entirely new one? In the case of an existing project, please explain the links between the two projects
A. Pasini and R. Clément are co-supervisors of Marvin Leria, a third year CENTURI PhD student working on the re-organization of cytoskeletal and planar polarity in Trichoplax epithelial cells during mechanical wound reparation. The proposed project will expand the knowledge and tools developed during the ongoing one all while addressing a much more physiologically relevant situation.
2 to 5 references related to the project
- Smith, C. L., Varoqueaux, F., Kittelmann, M., Azzam, R. N., Cooper, B., Winters, C. A., Eitel, M., Fasshauer, D.
and Reese, T. S. (2014). Novel cell types, neurosecretory cells, and body plan of the early-diverging metazoan
Trichoplax adhaerens. Current biology: CB, 24(14), 1565–1572. https://doi.org/10.1016/j.cub.2014.05.046 - Srivastava, M., Begovic, E., Chapman, J., Putnam, N.H., Hellsten, U., Ka- washima, T., Kuo, A., Mitros, T.,
Salamov, A., Carpenter, M.L., et al. (2008). The Trichoplax genome and the nature of placozoans. Nature 454,
955–960. https://doi.org/10.1038/nature07191. - Armon, S., Bull, M.S., Aranda-Diaz, A and Prakash, M. (2018) Ultrafast epithelial contractions provide insights
into contraction speed limits and tissue integrity. Proc Natl Acad Sci U S A. 2018 Oct 30;115(44):E10333-
E10341. doi: 10.1073/pnas.1802934115. - Varoqueaux, F., Williams, E. A., Grandemange, S., Truscello, L., Kamm, K., Schierwater, B., Jékely, G., &
Fasshauer, D. (2018). High Cell Diversity and Complex Peptidergic Signaling Underlie Placozoan
Behavior. Current biology : CB, 28(21), 3495–3501.e2. https://doi.org/10.1016/j.cub.2018.08.067 - Najle, S.R., Grau-Bové, X., Elek, A., Navarrete, C., Cianferoni, D., Chiva, C., Cañas-Armenteros, D.,
Mallabiabarrena, A., Kamm, K., Sabidó, E., Gruber-Vodicka, H., Schierwater, B., Serrano, L., and Sebé-Pedrós
A.(2023) Stepwise emergence of the neuronal gene expression program in early animal evolution.
Cell 186 (21):4676-4693.e29. doi: 10.1016/j.cell.2023.08.027
Two main publications from each PI over the last 5 years
- Chuyen A., Daian F., Pasini. A* and Kodjabachian L.* (2021) A live-imaging protocol to track cell movement in the Xenopus embryo. STAR Protoc 2 (100928) *Equal contribution
- Chuyen A., Rulquin C., Daian F., Thomé V., Clément R., Kodjabachian L. and Pasini. A. (2021) The SCF/KIT pathway implements self-organised epithelial patterning. Dev. Cell 56 (6), 795-810.e7
- Kong, W., Loison, O., Shivakumar, P., Chan, E.H., Saadaoui, M., Collinet, C., Lenne, P-F. and R. Clément. (2019)
Experimental validation of force inference in epithelia from cell to tissue scale. Scientific Reports - P. Roudot et al., (2023) u-track3D: Measuring, navigating, and validating dense particle trajectories in three dimensions Cell Reports Methods, doi: 10.1016/j.crmeth.2023.100655.
- J. Vanaret, V. Dupuis, P.-F. Lenne, F. J. Richard, S. Tlili, and P. Roudot, (2023) A detector-independent quality score for cell segmentation without ground truth in 3D live fluorescence microscopy. (Preliminary Acceptance) IEEE Journal of Selected Topics in Quantum Electronics, https://hal.archives-ouvertes.fr/hal-03923509
Project's illustrating image