PHD2020-21
Deciphering the cellular dynamics underlying macrophage network homeostasis
Host laboratory and collaborators
Marc Bajénoff / CIML/ bajenoff@ciml.univ-mrs.fr
Jean-François Rupprecht / CPT / jean-francois.rupprecht@cpt.univ-mrs.fr
Abstract
Macrophages are immune cells whose function is to destroy pathogens (eg. microbes or cancer cells) and ensure tissue homeostasis In particular, macrophages cover the surface of our epidermis through a paving network that is self-maintained, with minimal contribution from blood circulating monocytes.
We recently developed a multicolor rainbow-like fluorescent fate mapping system to investigate the dynamics of macrophage death and renewal. Our experimental imaging system revealed that the epidermal macrophage network is constituted by adjacent proliferative units composed of distinct macrophage cell populations called clones (See figure 1 below).
It is generally assumed that, when a macrophage dies, any of its neighbouring cell can divide to fill up the gap left within the defense network.
However, the theoretical results that we obtained based on this hypothesis do not match with the evolution in the clonal cell population levels that we experimentally observe.
We therefore hypothesize the existence of macrophage stem cells, which would be equally distributed among clones, thus regulating their population level. Hereby, we will investigate this hypothesis by combining theoretical simulations (J.F Rupprecht) and biological experimentations (M. Bajénoff).
Keywords
Macrophages, stem cell, computational simulation, imaging, fate mapping
Objectives
The overarching goal of this proposal is to decipher the cellular mechanisms underlying self-maintenance of macrophage networks across several tissues (skin, liver, brain). We ambition to create a paradigm shift demonstrating that macrophage rely on stem cells to self maintain over time, a strategy that is abundantly used by other cell types such as epithelia and other immune cell types.
Preliminary simulations suggest that only the one clonal cell population that is the most abundant initially will eventually survive after a sufficiently large number of cell divisions. Since this scenario does not correspond to current results obtained on aged mice, we would like to propose an alternative model in which individual clones would be continuously renewed by local macrophage stem cells that remains to be identified.
Proposed approach (experimental / theoretical / computational)
The approach is straightforward:
- Generation of mice in which tissue-resident macrophage will randomly acquire a color during their maturation and keep it for their entire lifetime (see Figure 1 below)
- Confocal microscopy analysis of tissue section and generation of Voronoi grids in which the number, colors and position of each macrophage will be recorded. This analysis will be done in young mice (starting point) and old mice (>12 months, end point).
- Computational simulation in which a random fraction of the macrophage pool observed in young animals will be replaced randomly and repeatedly over time in order to mimic natural turnover.
- Determine if the simulated network matches the experimental data obtained in old animals.
- Search for macrophage stem cells using specific mouse models allowing tracking and isolation of dividing macrophage (Fucci mice) or stem cell (mouse models reporting Wnt signalling activity).
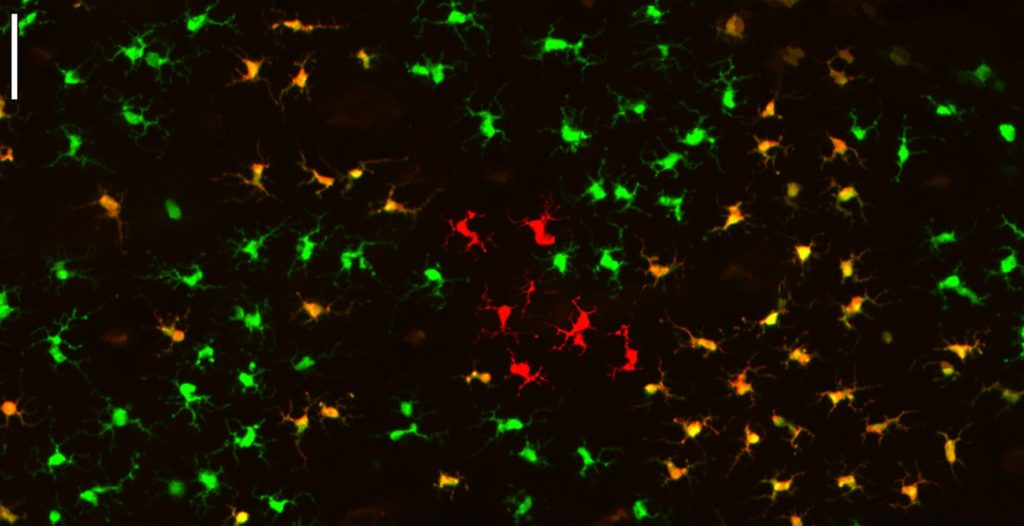 Figure 1: Multicolor fate mapping of epidermal macrophages
Figure 1: Multicolor fate mapping of epidermal macrophages
(left) Confocal image of a piece of the network of epidermal macrophage harvested from the ear of a Langherin-Cre+ Ubow+/+ mice. In these mice, all epidermal macrophages randomly express two copies of YFP (green), two copies of CFP (red) or one copy of each fluorescent protein (orange). Yet, note how these macrophage associate in monocolored clusters resulting from the clonal proliferation of few macrophage.
From J Exp Med. 2013 Aug 26;210(9):1657-64
Interdisciplinarity
This proposal is interdisciplinary because it involves two teams that are expert in biology and theoretical/computational simulation. From a practical point of view, the biological partner will provide raw data to the computational team which will then provide models and hypotheses to the biological partner who will design new experiments to test these hypotheses. This back and forth discussion represents the foundation of this proposal and this project could not exist without it. The two teams are located on the Luminy Campus, which will ease the interactions.
Expected profile
Developmental biologist or immunologist interested in computational biology.
