PHD2024-17
EndoEmbryo: toward a data-driven understanding of how endo/exocytic fluxes shape embryo morphogenesis
Host laboratory and collaborators
Phillipe Roudot, Institut Fresnel philippe.roudot@univ-amu.fr
Claudio Collinet, IBDM Claudio.collinet@univ-amu.fr
Abstract
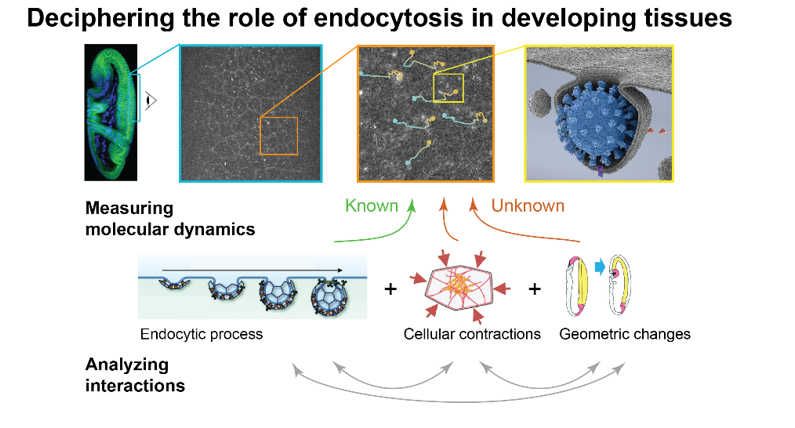
The ability of cells and tissues to change shape during development relies on patterns of molecules at the cell surface, such as adhesion molecules and signaling receptors, that anchor and regulate cell contractions. These processes emerge from the collective dynamics of hundred-to-thousand agents (molecules, polymers, molecular motors, cellular organelles etc.) which can be observed in vivo through time-lapse fluorescence microscopy. However, the stochastic nature of the interactions amongst the agents challenges our understanding of how cellular functions emerge from their collective dynamics. This project proposal aims at using image time series to probe the non-linear link between the recruitment of thousands of interacting molecules, the robust molecular patterning that emerges and the tight orchestration of contractile events.
Specifically, the project will focus on a data-driven study of endo/exocytosis (E/E), a process that endows the cell with a capacity to exchange and redistribute molecules at the cell surface. Through the collective dynamics of hundreds of membrane intracellular organelles, E/E allows cells to establish and maintain spatial patterns of adhesion and signaling molecules which, in turn, regulate cellular contractility. How E/E is regulated and in turn regulates tissue morphogenesis is still largely unknown due to the challenge of tracking and measuring the dynamics of hundred-to-thousands organelles in noisy images. This project will be based our recent work on non-supervised multiple object tracking and large language modeling of intracellular dynamics to drive the calibration of 3D confocal imaging experiment and optimize the quantification of molecular events. By parameterizing both the image formation process and the multiscale biological interactions, these approaches will help decipher how cells recruit, deplete, and translocate molecules to maintain spatial patterns that are crucial to shape formation.
Keywords
Tissue morphogenesis, collective dynamics, intracellular trafficking, multiple object tracking, large language model.
Objectives
The first objective will consist in developing an integrated computational and experimental framework to measure the spatial distribution of E/E fluxes of cell adhesion molecules and signalling receptors (GPCRs) in two distinct contexts of morphogenesis in the fly embryo. This framework will then be applied to: 1) characterize any heterogeneity in endocytic particle dynamics relative to cell adhesion molecules and GPCRs surface distributions, and 2) to detect any temporal hierarchy between endocytic events and patterns and dynamics of actomyosin contractility during morphogenesis.
Proposed approach (experimental / theoretical / computational)
Living embryos will be imaged by spinning disk confocal microscopy to track endocytic events in 3D over time. Markers of endocytosis (Clathrin, AP-2) and of exocytosis (Sec5, Sec15) will be imaged together with endocytic cargo molecules (E-Cad, Integrins, and GPCRs) in two tissues undergoing different types of morphogenetic events. Actomyosin contractility and cell shape will be monitored to correlate endocytic events to cell and tissue morphogenesis. The quantification of endocytic turn over is based on a stochastic modeling of both the biophysical process of clathrin-coated pits formation and the noisy nature of fluorescence imaging. In order to compensate for the multi-scale nature of epithelial motion that challenges the tracking of an object from one frame to the next, we will propose a new approach that enables the decrease of apparent motion magnitude by increasing the framerate/SNR ratio. This regime of acquisition requires the registration of many false targets in combination with our targets of interest (the endocytic pits). Our preliminary work shows that attention-based large language model significantly push the boundaries of existing methods in those scenarios. This approach will be completed with a stochastic modeling and inference of the temporal hierarchy between molecular events (endocytosis, adhesion recruitment, actomyosin mechanics).
Interdisciplinarity
The PhD student will be co-hosted by the Roudot group (Inst. Fresnel, an engineering laboratory) and the Lecuit group (IBDM, a developmental biology institute). As such, tShey will be deeply involved in an interdisciplinary conversation with different expertise in computer vision, cellular biology, biochemistry and microscopy. First, the optimal alignment of techniques must be explored to optimize the measure of molecular events at scale: setting the blueprint for optimal fluorescent labelling, illumination level and sampling adjustment as well as algorithm selection and parameterization. Second, the design of the motion model must be chosen as a trade-off between physical accuracy and computational feasibility, requiring strong mentoring in complex systems physics and computer vision. Similarly, the interpretation of biological measurements across scales to understand signaling pathways and setting the next experiment also requires expertise in cellular and system biology. Finally, the development of perennial tools to study those complex 3D systems involves significant training in numerical methods for stochastic inferences.
Expected profile
The PhD student will have a formal training in applied mathematics, computer science or biophysics with a keen interest in cell biology and the study of complex systems.
Is this project the continuation of an existing project or an entirely new one? In the case of an existing project, please explain the links between the two projects
It is a follow up study on a preliminary benchmark on tracking approach for the developing embryo.
2 to 5 references related to the project
- R. Levayer, A. Pelissier-Monier, and T. Lecuit, “Spatial regulation of Dia and Myosin-II by RhoGEF2 controls initiation of E-cadherin endocytosis during epithelial morphogenesis,” Nat Cell Biol, vol. 13, no. 5, Art. no. 5, May 2011, doi: 10.1038/ncb2224.
- R. Levayer and T. Lecuit, “Oscillation and Polarity of E-Cadherin Asymmetries Control Actomyosin Flow Patterns during Morphogenesis,” Developmental Cell, vol. 26, no. 2, pp. 162–175, Jul. 2013, doi: 10.1016/j.devcel.2013.06.020.
- Sigismund, S., Lanzetti, L., Scita, G. & Di Fiore, P. P. Endocytosis in the context-dependent regulation of individual and collective cell properties. Nat. Rev. Mol. Cell Biol. 22, 625–643 (2021).
- K. Granström, M. Fatemi, and L. Svensson, “Poisson Multi-Bernoulli Mixture Conjugate Prior for Multiple Extended Target Filtering,” IEEE Transactions on Aerospace and Electronic Systems, vol. 56, no. 1, pp. 208–225, Feb. 2020, doi: 10.1109/TAES.2019.2920220.
- Bailles A., Collinet C., Philippe J-M., Lenne P-F., Munro E., Lecuit T. “Genetic induction and mechanochemical propagation of a morphogenetic wave” Nature 2019 Aug;572(7770):467-473. doi: 10.1038/s41586-019-1492-9.
Two main publications from each PI over the last 5 years
- P. Roudot et al., (2023) u-track3D: Measuring, navigating, and validating dense particle trajectories in three dimensions. Cell Reports Methods, in press. doi: 10.1016/j.crmeth.2023.100655.
- J. Vanaret, V. Dupuis, P.-F. Lenne, F. J. Richard, S. Tlili, and P. Roudot, (2023) A detector-independent quality score for cell segmentation without ground truth in 3D live fluorescence microscopy. IEEE Journal of Selected Topics in Quantum Electronics, Jan. 2023, https://hal.archives-ouvertes.fr/hal-03923509
- Collinet C. and Lecuit T. “Programmed and self-organized flow of information during morphogenesis.” Nat Rev Mol Cell Biol. 2021 Apr;22(4):245-265. doi: 10.1038/s41580-020-00318-6.
- Bailles A., Collinet C., Philippe J-M., Lenne P-F., Munro E., Lecuit T. “Genetic induction and mechanochemical propagation of a morphogenetic wave” Nature 2019 Aug;572(7770):467-473. doi: 10.1038/s41586-019-1492-9.
Project's illustrating video and image