PHD2024-10
Force-watch: a novel LCD based tool to measure feeble forces generated by immune cells
Host laboratory and collaborators
Kheya SENGUPTA, CINaM kheya.sengupta@cnrs.fr
Aurélien Dumètre, LAI aurelien.dumetre@univ-amu.fr
Abstract
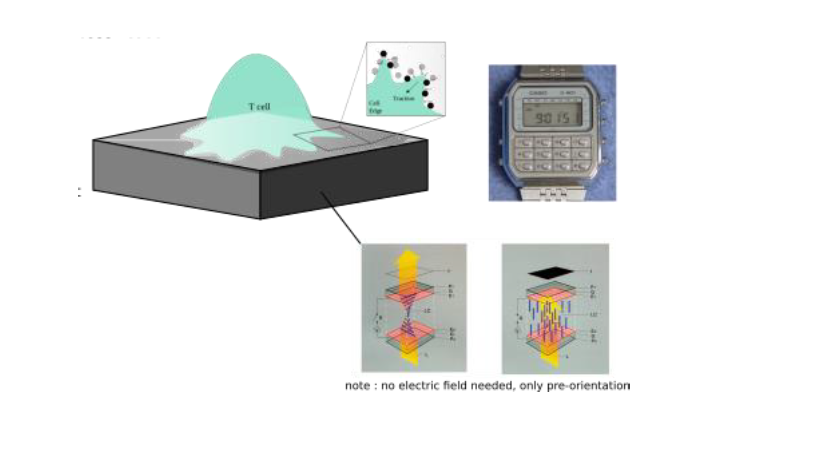
An important missing piece in our understanding of immune cells concerns reliable measurement of the force they generate to detect and eliminate abnormal or foreign cells. While traction force microscopy (TFM) is now fairly well established for measurements of forces in tissue forming cells, new tools are desperately needed to enable measurement of feeble traction forces, such as those exerted by leucocytes under physiologically relevant conditions. In addition, simultaneous high resolution optical microscopy to reveal the dynamics of the different molecular players in force generation is desirable. This project aims at developing a new tool to measure sub-nano-Newton forces applied by leucocytes such as phagocytes (which use forces to attach, engulf and deform targets to break them) and lymphocytes (which punch target cells to induce their death), especially during early contact with a ligand presenting cell surface or microbes. The “force-watch” will exploit the technology used in a basic liquid crystal display (LCD), such as those found on wrist-watches where light passing through an anisotropic medium and under crossed-polarizers encodes distortions as shades of grey. These distortions, as in conventional TFM, can be used to infer forces. We plan to use machine learning (ML) based image analysis to bypass the need for solving complex equations both to infer distortions from light patterns and forces from distortion patterns. Using light in this way to measure continuous molecular distortions in the medium, without imaging extrinsic microscopic markers, will allow us to overcome problems of discrete measurements.
Keywords
Traction force microscopy, Leucocytes, Optical anisotropy, Imaging
Objectives
The overall objective of this project is to measure the sub-nano Newton local forces generated by immune cells for their physiological functions, which appear to be tightly controlled in terms of spatial and temporal patterns. Two main limitations of current force measurement strategies are that the forces are reported at discreet points, and that being dependent on measuring displacements with finite resolution, it is difficult to measure feeble forces. The aim of this project is to overcome these limitations by quantifying forces from continuous distortions of an anisotropic elastomer, read out using technology similar that used for LCD screens. This novel device will be used to measure forces exerted by immune cells such as macrophages and lymphocytes.
Proposed approach (experimental / theoretical / computational)
Immune cells are highly dynamic, constantly changing shape, migrating, and intermittently interacting with other cells. Major examples include macrophages which need to apply forces to their targets to attach, engulf, break, and digest them [1], and T lymphocytes, which employ mechanical forces to identify pathogenic antigens on the surface of a target cell and to kill it [2]. The spatio-temporal pattern of the applied traction force offers crucial insights into cell behaviour. TFM aims to measure these patterns on engineered substrates with well-defined elastic properties, utilizing deformation information extracted from substrate images [2]. We seek to improve traditional TFM by developing a novel experimental tool to measure feeble forces by-passing current microscopic marker-based detection, that is already mastered in the team [3]. Instead, detection will be based on interaction of polarized light with a deformable anisotropic medium whose distortion in response to cell-applied forces will be optically detected. For that, we plan use deformable polymer-based liquid crystal elastomers, that will be preordered magnetically, and will, like an LCD watch, reveal deformation of the substrate upon changes in orientation, without the need of any extrinsic marker. ML, which is currently being explored for conventional TFM [4], will be used to bypass onerous calculations. The optical strategy of using polarized light also frees up a fluorescence channel for bio-labelling the cells to simultaneously reveal membrane and cytoskeletal structures in real-time. The tasks to be undertaken will be:
- Set-up Force-watch, observe large macrophage generated distortions
- Calibrate and use ML to infer forces from distortions, cross-validate using conventional TFM.
- Optimize to measure feeble forces and apply to both macrophage and T lymphocytes
Interdisciplinarity
In this collaboration, CINaM (Physics) will bring expertise on material science and the techniques of micro and nano-engineering; LAI (Biology) will bring expertise on immunology, especially on microbe-phagocyte interactions. Neither partner can hope to achieve the goals alone. LAI lacks relevant expertise in materials and patterning and CINaM lacks competence to access important biological tools. Traditional TFM approaches will be supported by Pierre-Henri Puech (LAI), coupling imaging techniques by Laurent Limozin (LAI) and Machine Learning tasks will be supported by Pierre Ronceray (CINaM), putting this project in the frame of a larger interdisciplinary and inter-lab effort to quantitatively approach forces and their consequences in immunological systems. In addition, we shall benefit from the technology platform of Centuri for help with fabrication as well as computational support that will be needed for quantifying the distortion and converting detected distortions to a measured force. We hope to use this PhD as a stepping-stone to being the newly developed tools to the Centuri community.
Expected profile
The candidate should have an academic background in physics/engineering or biophysics. Candidates with previous experience in optical microscopy will be given preference. Reasonable competence in computer programming is expected. We look for highly motivated candidates willing to do experimental and computational work at the interface of physics and biology, in two interdisciplinary laboratories gathering physicists, biologists and medical doctors.
Is this project the continuation of an existing project or an entirely new one? In the case of an existing project, please explain the links between the two projects
Entirely new. However, “Force-watch” fits into broader on-going collaborative effort between LAI and CINaM to overcome current limits of traction force microscopy and of biophysical techniques applied to single cells studies, in particular in immunology.
2 to 5 references related to the project
- Dynamics of Toxoplasma gondii oocyst phagocytosis by macrophages. Ndao O., Puech P.H., Bérard C., Limozin L., RabhiS., Azas N., Dubey J.P., Dumètre A. Frontiers in Cellular and Infection Microbiology, 2020, 10:207.doi.org/10.3389/fcimb.2020.00207
- May the force be with your (immune) cells: an introduction to traction force microscopy in Immunology. F. Mustapha, K. Sengupta, P-H. Puech. Frontiers in Immunology (2022). DOI: 10.3389/fimmu.2022.898558
- Protocol for measuring weak cellular traction forces using well-controlled ultra-soft polyacrylamide gels, F. Mustapha, K. Sengupta, P-H. Puech. STAR Protocols (2022). DOI: 10.1016/j.xpro.2022.101133[4] Enhancing robustness, precision, and speed of traction force microscopy with machine learning, F. S. Kratz, L. Möllerherm, J. Kierfeld, J. Biophysical Journal (2023). DOI: 10.1016/j.bpj.2023.07.025
Two main publications from each PI over the last 5 years
KHEYA SENGUPTA
- Ligand Nanocluster Array Enables AI Based Detection of Hidden Features in T-Cell Architecture, A. Nassereddine, A. Abdelrahman, … K. Sengupta Nano Lett. 21(13):5606-5613 (2021).
- Biphasic mechanosensitivity of TCR mediated adhesion of T lymphocytes. A. Wahl, C. Dinet, P. Dillard, P-H. Puech, L. Limozin, and K. Sengupta. PNAS 116 (13) 5908-5913 (2019).
AURELIEN DUMETRE
- Detection, fate, and transport of the biohazardous agent Toxoplasma gondii in soil water systems: influence of soil physicochemical properties, water chemistry, and surfactant. Kinsey E.N., Korte C., Gouasmia S., L’Ollivier C., Dubey J.P., Dumètre A., Darnault C.J.G. Environmental Microbiology Reports (2023) 15:596-613. doi.org/10.1111/1758-2229.13204
- Structure, composition, and roles of the Toxoplasma gondii oocyst and sporocyst walls. Freppel W., Ferguson D.J.P., Shapiro K., Dubey J.P., Puech P.H., Dumètre A. The Cell Surface (2019) 5:100016. doi.org/10.1016/j.tcsw.2018.100016
Project's illustrating image