PHD2024-07
Salmonella facing neutrophils: Characterizing HOCl stress response and resulting population dynamics
Host laboratory and collaborators
Maxence Vincent, Laboratoire de Chimie Bactérienne mvincent@imm.cnrs.fr
Florence Bansept, Laboratoire de Chimie Bactérienne Florence.bansept@univ-amu.fr
Abstract
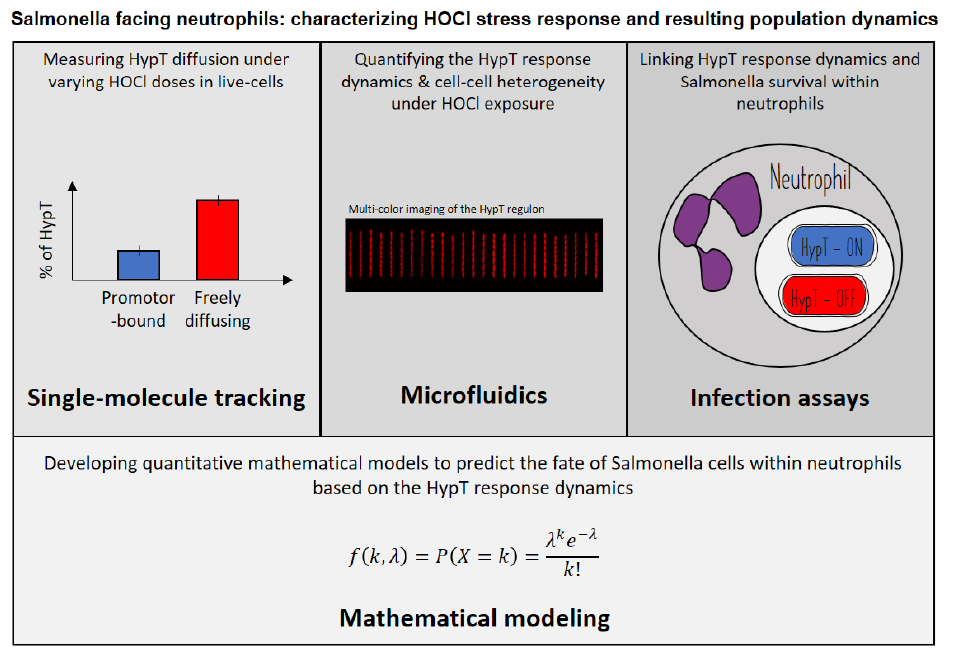
Salmonella is a pathogenic bacterium that poses a significant threat to public health. During infections, neutrophils secrete hypochlorous acid (HOCl) as a potent weapon to eliminate pathogens. This project aims to investigate the population dynamics of Salmonella during HOCl stress, with a primary focus on the transcription factor HypT, the key regulator of the HOCl stress response. The project will adopt an interdisciplinary approach that includes the utilization of cutting-edge microscopy techniques to visualize the HypT response in live Salmonella cells at both the single-cell and single-molecule levels. Additionally, mathematical models of explicitly interacting Salmonella and neutrophils populations will be developed. By accounting for the physical properties of the involved molecules, they will allow to predict the fates of these populations in host-pathogen infection experiments. By unveiling the molecular processes that underlie Salmonella's adaptation to the immune oxidative burst, we will pave the way to the understanding of a key aspect of host-pathogen coevolution.
Keywords
Population dynamics – Oxidative Stress – Host-pathogen Interaction – Salmonella Typhimurium – Microfluidics –Single-particle tracking – Mathematical Modelling
Objectives
The proposed project can be divided into four objectives:
I. Deciphering the kinetics of HypT activation at thesingle-molecule level within live Salmonella cells.
II. Quantifying the regulatory dynamics of HypT-regulatedgenes at the single-cell and the population levels.
III. Monitoring HypT activation during Salmonella-neutrophilinteractions and correlating it with Salmonella survival within the host.
IV: Developing quantitative models basedon the collected experimental data to predict the fate of Salmonella cells within neutrophils.
Proposed approach (experimental / theoretical / computational)
The project is experimentally based on cutting-edge imaging techniques to investigate the HypT response to HOCl stress. Live-cell single-particle tracking of HypT, multi-color microfluidics-based imaging, and in-host time-lapse microscopy will provide unprecedented insights into (i) HypT's diffusive behavior, (ii) the regulatory dynamics of the HypT regulon under HOCl treatments, and (iii) HypT's role in Salmonella's adaptation to the immune oxidative burst. The data generated by these experiments will necessitate computational processing, including the use of single-particle localization and tracking algorithms, single-cell segmentation methods, and correlative analyses. In parallel with this experimental workflow, we will develop quantitative mathematical models to predict the fate of Salmonella cells within neutrophils, thus establishing a link between the HypT response and Salmonella's fate during host infection.
Interdisciplinarity
This project embodies an interdisciplinary approach, combining genetic engineering of pathogenic bacteria, state-of-the-art imaging methods, and mathematical modeling. While the project is ambitious, it benefits from the expertise of two PIs with complementary skills, fostering interdisciplinary synergy. PI.1 (MV), a microbiologist, became expert with live-cell single-molecule tracking and microfluidic-based microscopy during his postdoctoral training to elucidate phenotypic heterogeneity's role in bacterial stress responses. Recently appointed as a CRCN at CNRS (October 2023), he aspires to apply similar approaches to host-infection-related questions. PI.2 (FB), a theoretical biologist specialized in the mathematical modelling of microbial communities, leads a CENTURI research group. She is an expert in within-host population dynamics, and related evolutionary questions. Overall, this collaboration seamlessly integrates experimental and theoretical approaches to unveil novel insights into Salmonella's response to immune defenses, fostering a holistic understanding of the topic.
Expected profile
The ideal PhD candidate for this interdisciplinary project should have a strong background in quantitative microbiology and bacterial stress responses. Proficiency in advanced imaging techniques is crucial. The candidate must possess robust quantitative and computational skills, including experience in coding, image analysis, and data processing. This project necessitates the ability to creatively integrate experimental and theoretical work. The student should be capable of conducting experimental research, while also effectively contextualizing their results within a theoretical framework. Given that the project requires the candidate to reflect on their project from both experimental and theoretical perspectives, excellent communication skills are essential for presenting complex scientific data to diverse audiences, including experimentalists and theorists. The PhD candidate should be enthusiastic about developing new methods of investigations in the field of host-pathogen interaction. Finally, a commitment to ethical research practices is fundamental.
Is this project the continuation of an existing project or an entirely new one? In the case of an existing project, please explain the links between the two projects
Entirely new one.
2 to 5 references related to the project
- Schürmann, N. et al. Myeloperoxidase targets oxidative host attacks to Salmonella and prevents collateral tissue damage. Nat Microbiol 2, 16268 (2017).
- Jo, I. et al. Structural basis for HOCl recognition and regulation mechanisms of HypT, a hypochlorite-specific transcriptional regulator. Proceedings of the National Academy of Sciences 116, 3740–3745 (2019).
- Drazic, A. et al. Methionine oxidation activates a transcription factor in response to oxidative stress. Proceedings of the National Academy of Sciences 110, 9493–9498 (2013).
- Gray, M. J., Wholey, W.-Y. & Jakob, U. Bacterial responses to reactive chlorine species. Annu Rev Microbiol 67, 141–160 (2013).
Two main publications from each PI over the last 5 years
Maxence VINCENT
Vincent, M. S. & Uphoff, S. Cellular heterogeneity in DNA alkylation repair increases population genetic plasticity. Nucleic Acids Research 49, 12320–12331 (2021).
Vincent, M. S. et al. Dynamic proton-dependent motors power type IX secretion and gliding motility in Flavobacterium. PLOS Biology 20, e3001443 (2022).
Florence Bansept
Bansept F, Schumann-Moor K, Diard M, Hardt WD, Slack E, Loverdo C. Enchained growth and cluster dislocation: A possible mechanism for microbiota homeostasis. PLOS Computational Biology. 2019;15(5) :e1006986.
Bansept F, Obeng N, Schulenburg H, Traulsen A. Modeling host-associating microbes under selection. ISME J. 2021 Jun ;p. 1–9.
Project's illustrating image