PHD2024-25
Videonanoscopy of the thymic immunological synapse during leukemogenesis
Host laboratory and collaborators
Arnauld SERGÉ, LAI arnauld.serge@univ-amu.fr
Magali IRLA, CIML irla@ciml.univ-mrs.fr
Abstract
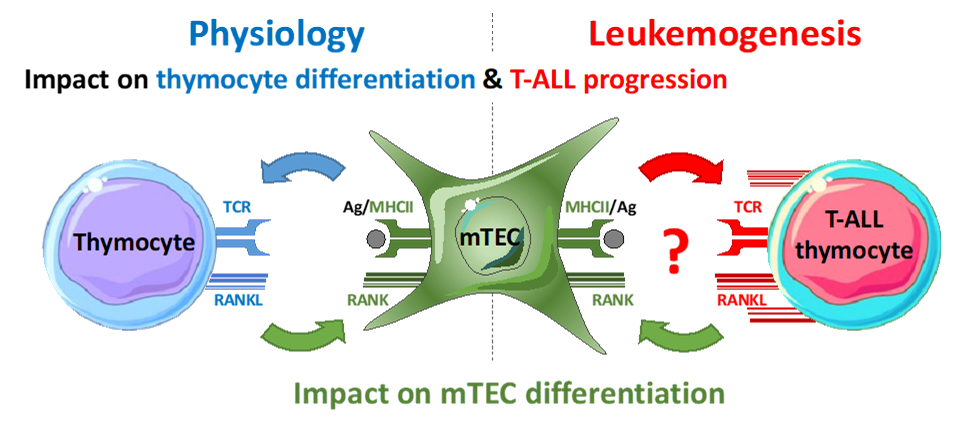
T-cell acute lymphoblastic leukemia (T-ALL) result from the malignant transformation of developing T cells. With a poor prognosis, they lead to relapse due to resistance to chemotherapy. Thymic epithelial cells (TEC) and leukemic cells stimulate each other by unknown signals. Our preliminary data show a gradual overexpression of the cytokine RANKL by leukemic cells in a spontaneous mouse model of T-ALL. Furthermore, the addition of an anti-RANKL blocking antibody to a coculture of leukemic cells and TECs expressing the RANK receptor inhibits leukemia growth. Although its role in leukemogenesis is unknown, the RANK/RANKL axis, involved in several tumors, is a promising therapeutic target. The challenge of this project is to study RANK/RANKL interactions between TECs and T-ALL cells by fast videonanoscopy, determining the spatiotemporal entanglement of the RANK and TCR pathways at the immunological synapse and their deregulation in T-ALLs.
Keywords
Single molecule tracking, super-resolution, videonanoscopy, diffusion, membrane dynamics, leukemia, immune synapse, T cell, medullary thymic epithelial cell, RANK/RANKL, image analysis
Objectives
Our hypothesis is that the signals exchanged between TECs and T-ALL cells play a key role in leukemogenesis. The objective is to determine the spatiotemporal organization of RANK and RANKL at the immunological synapse and to evaluate its impact on leukemogenesis. We will attempt to disrupt these dynamics using an anti-RANKL blocking antibody, to test new therapeutic strategies for T-ALL.
Proposed approach (experimental / theoretical / computational)
In the context of this biophysical project, we will use fast videonanoscopy to measure diffusion within cell membranes. Ligand/receptor interactions will be studied using physiological and leukemic cell models and innovative tools for the analysis of molecular trajectories, reconstructed by a proprietary algorithm, Multi-Target Tracing. We will develop a pipeline of Matlab functions dedicated to the characterization of these measurements. We will identify ligand/receptor interactions at contacts between TECs and T-ALL cells. We will extend our measurements to the simultaneous tracking of TCR, MHCII, RANK and RANKL, and then to primary normal or leukemic T cells, from healthy donors or patients.
Interdisciplinarity
This interdisciplinary project targets the interactions between leukemic and stromal cells in the thymus by single-molecule tracking. The aim of this PhD project, at the interface between immunology, biophysics, and image analysis, is to characterization molecular dynamics at contacts between TECs and T-ALL cells. The project addresses on one side the Brownian motion of membrane receptors and its repercussions on the organization of cell contacts, with a need to develop innovative imaging techniques and analysis software. On the other side, it concerns the biology of cellular and molecular interactions during leukemogenesis, with a need for tools to improve single molecule labeling.
Expected profile
We are seeking for an interdisciplinary profile in biophysics, with a background in immunology or cancer. Knowledge in cell and molecular biology would be an asset. Concerning the biophysical side of the project, we are looking for a candidate with good experience in programming, in Matlab or equivalent, for image analysis and basic knowledge in optics. Background in statistics would also be appreciated. Language: good level in English, French would be a plus.
Is this project the continuation of an existing project or an entirely new one? In the case of an existing project, please explain the links between the two project
We have already started to investigate the dynamics of the immune synapse. We are setting up the protocols for TECs and T-ALL cells labeling and videonanoscopy measurements. We are further developing the analysis codes to extend our analyses to any number of molecular populations, in terms of colocalizations and combined distribution of relevant parameters.
2 to 5 references related to the project
- Lopes N, Vachon H, Marie J, Irla M. Administration of RANKL boosts thymic regeneration upon bone marrow transplantation. EMBO Mol Med. 2017
- Lopes N, Sergé A, Ferrier P, Irla M. Thymic Crosstalk Coordinates Medulla Organization and T-Cell Tolerance Induction. Front Immunol (2015)
- Ahern, E., Smyth, M.J., Dougall, W.C. et al. Roles of the RANKL–RANK axis in antitumour immunity — implications for therapy. Nat Rev Clin Oncol 2018
- Sergé, A., Bertaux, N., Rigneault, H., Marguet, D. Dynamic multiple-target tracing to probe spatiotemporal cartography of cell membranes. Nature Methods 2008
- Rouger, V., Bertaux, N., Trombik, T., Mailfert, S., Billaudeau, C., Marguet, D., Sergé, A. Mapping molecular diffusion in the plasma membrane by Multiple-Target Tracing (MTT). Journal of visualized experiments 2012
Two main publications from each PI over the last 5 year
Arnauld Sergé
- Gorshkova O, Cappaï J, Maillot L, Sergé A*. Analyzing normal and disrupted leukemic stem cell adhesion to bone marrow stromal cells by single-molecule tracking nanoscopy. J Cell Sci. 2021 *Corresponding
- Charaix J, Borelli A, Santamaria JC, Chasson L, Giraud M, Sergé A, Irla M. Recirculating Foxp3+ regulatory T cells are restimulated in the thymus under Aire control. Cell Mol Life Sci. 2022
Magali Irla
- Lopes N, Boucherit N, Santamaria JC, Provin N, Charaix J, Ferrier P, Giraud M, Irla M. Thymocytes trigger self-antigen-controlling pathways in immature medullary thymic epithelial stages. Elife. 2022 Feb 21;11:e69982.
- Lopes N, Charaix J, Cédile O, Sergé A, Irla M*. Lymphotoxin alpha fine-tunes T cell clonal deletion by regulating thymic entry of antigen-presenting cells. Nature Communications 2018 *Corresponding
Project's illustrating image