PHD2024-16
Mapping non-equilibrium fluctuations in the developing fly embryo
Host laboratory and collaborators
Emily Gehrels, CINaM emily.gehrels@univ-amu.fr
Pierre Ronceray, CINaM pierre.ronceray@univ-amu.fr
Abstract
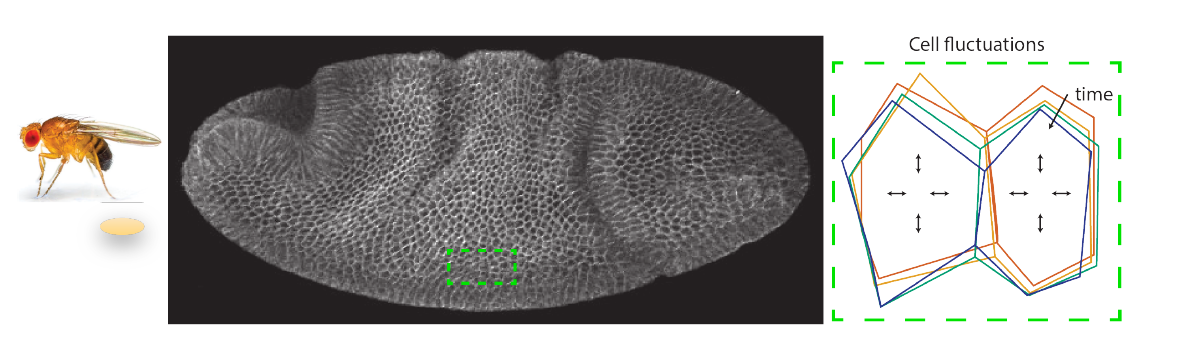
Cold-blooded animals, which cannot regulate their temperature, develop faster in hot environments than in cold ones – but they achieve the same final form. This simple observation is actually very surprising since the rates of the many chemical reactions and transport phenomena involved in development do not depend on temperature in the same way, and so well-understood thermal effects must be actively compensated by yet-unknown regulation mechanisms. Can we map the contributions of these active mechanisms in space and time by measuring metabolic rates and non-equilibrium fluctuations in live organisms?
The student will tackle this problem by investigating the temperature-dependence of Drosophila embryo development, a model organism uniquely suited to decoupling the effects of thermal and actively-regulated processes. By combining liveimaging of the embryo under different, controlled conditions with modern analytic tools to quantify non-equilibrium fluctuations in a non-intrusive way, we will establish a first multi-scale activity map of the developing embryo. This will open a new window towards understanding the robustness of life to environmental perturbations.
Keywords
Tissue dynamics, Embryonic development, Non-equilibrium processes, inference
Objectives
Our objectives are to: (1) correlate cell-scale fluctuations to developmental speed by quantifying cell shape dynamics at different temperatures (and thus different developmental speeds); (2) decouple energy-consuming contributions to cell fluctuations from thermal ones by independently tuning metabolic pathways; and (3) establish the first multi-scale activity map of the developing embryo by directly extracting non-equilibrium contributions to fluctuations.
Proposed approach (experimental / theoretical / computational)
Experimentally, the project will utilize confocal microscopy to acquire cell- and tissue-scale time series of Drosophila embryos during development. These experiments will be performed at different temperatures and, independently, under exposure to drugs that affect different steps in the metabolic pathway, enabling us to change the rate of energy consumption. We will verify the effects of the drugs using existing readouts for metabolic activity at the cell and organism scale.
Once the data are acquired under these different conditions, we will use a combination of existing and novel data analysis techniques to extract quantitative information about the system. Image analysis software such as FIJI will be used for cell segmentation and tracking to extract cellular and subcellular dynamics. In parallel, we will implement and adapt recentlydeveloped tools for extracting the relative contributions of thermal and non-equilibrium processes to these dynamics.
Interdisciplinarity
This ambitious yet feasible PhD project bridges between EG’s expertise on Drosophila embryo manipulation and imaging, and PR’s expertise on non-equilibrium stochastic processes and data analysis. It is set at the interface between experimental developmental biology, bioenergetics and statistical mechanics, and will combine advanced microscopy with cutting-edge data analysis methods. This project is thus highly interdisciplinary both in its methods and in its goals.
Expected profile
We expect the student to have previous experimental lab experience, preferably including some experience with microscopy. The student should have a background in experimental physics of biological and/or soft matter systems, as well as proficiency with statistical physics tools (fluctuation-dissipation relation, Brownian dynamics…). Prior experience in image analysis (FIJI, Trackmate) and programming in Python would be a plus.
Is this project the continuation of an existing project or an entirely new one? In the case of an existing project, please explain the links between the two projects
This is an entirely new project.
2 to 5 references related to the project
- AL-Saffar, Grainger & Aldrich. Effects of constant and fluctuating temperature on development from egg to adult of Drosophila melanogaster (Meigen). Biol Environ 95B, 119–122 (1995).
- Crapse, et al. Evaluating the Arrhenius equation for developmental processes. Mol Syst Biol 17, e9895 (2021).
- Klepsatel, Wildridge & Gáliková. Temperature induces changes in Drosophila energy stores. Sci Rep 9, 5239 (2019).
- Kim, Pochitaloff, Stooke-Vaughan & Campas. Embryonic tissues as active foams. Nat Phys 17, 859-866 (2021).
- Muenker, Knotz, Krüger & Betz. Onsager regression characterizes living systems in passive measurements. bioRxiv doi: 10.1101/2022.05.15.491928 (2023).
Two main publications from each PI over the last 5 years
E. Gehrels
- Gehrels, E.W.*, Chakrabortty, B.*, Perrin, M.-E., Merkel, M., Lecuit, T. Curvature gradient drives polarized flow in the
Drosophila embryo. PNAS 120 (6), e2214205120 (2023). - Bailles, A.*, Gehrels, E.W.*, Lecuit, T. Mechanochemical principles of spatial and temporal patterns in cells and tissues.
Annual Review of Cell and Developmental Biology 38, 321-347 (2022).
P. Ronceray
- Frishman, A.*, Ronceray, P*. Learning force fields from stochastic trajectories. Physical Review X 10 (2), 021009 (2020).
- Ronceray, P. Two steps forward – and one step back? Measuring fluctuation-dissipation breakdown from fluctuations
only. Journal Club for Condensed Matter Physics 2 (July 2023).
Project's illustrating image