PHD2024-04
Multiscale killer-target dynamics in an immunotherapy assay
Host laboratory and collaborators
Laurent Limozin, LAI laurent.limozin@univ-amu.fr
Nicolas Levernier, CINAM nicolas.LEVERNIER@univ-amu.fr
Abstract
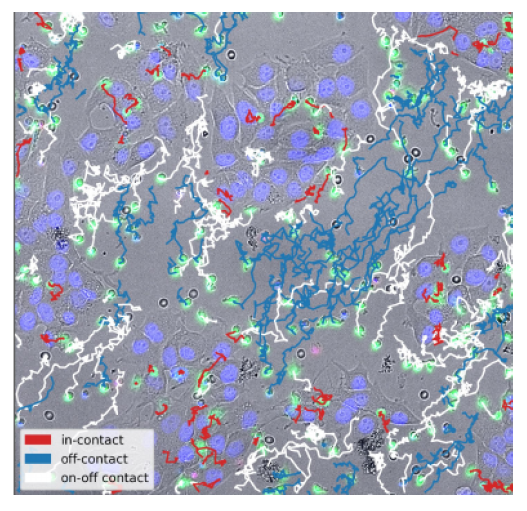
Immunotherapy holds great promises for fighting cancer, but the understanding of fundamental mechanisms of action of existing or potential treatments is often lacking, therefore limiting broader or more efficient applications. To address that, quantitative in vitro assays for testing and modeling phenomena are central. We developed a quasi 2D microscopy assay where cancer cells and human immune cells are co-cultured in presence of original antibodies forming molecular bridge between the two cell types. Using a powerful home-made image analysis software, cells can be followed during their interactions but we lack now a theoretical approach to model these complex phenomena. This will help to optimize new molecules for maximizing immune cell recruitment and target cancer cell killing, while minimizing drug amounts and side effects.
Keywords
Statistical physics; first-passage time; immunological synapse; predator-prey dynamics; bispecific antibody; molecular and cellular interactions
Objectives
The goal of the project is to develop a theoretical framework to analyze and interpret the dynamics of interactions between immune and target cells. A multiscale modeling would encompass the localisation and binding of antibodies, the threshold for cells response as well as the population spatio-temporal dynamics. It will rely on analytical tools and numerical simulations, in close comparison with the experiments.
Proposed approach (experimental / theoretical / computational)
The different tasks of the project will involve: (1) design numerical simulations to test and reproduce experimental data, using Langevin or more general stochastic dynamics (2) define a minimal set of parameters to describe natural and antibody-induced behaviour (kinetic parameters, effective interactions, etc.) (3) Adapt analysis to new image contents and be force of proposition for new experimental measurements (4) propose a functional back and forth pipeline between experiment and theory to accelerate the development of new diagnosis and molecules.The main dataset concerns bispecific mediated killing of tumour cell lines by Natural-Killer cells in videomicroscopy. Optional: It is complemented by higher resolution data of synapse formation on mimetic targets. Both datasets will be further extended within an on-going collaborative project lead by L. Limozin.
Interdisciplinarity
The project is interdisciplinary because original physics and computational tools will be developed to address quantitatively a biological system. Additionally , the analysis and modeling will be performed hand in hand with new experimental developments in LAI. Finally, the system is aimed to provide interesting physics as well as paving the way for more rational and efficient approaches in drug design.
Expected profile
The student will have preferentially a degree in physics or biophysics, ideally with an experience in quantitative biology modeling. It should have a strong experience in the study of stochastic dynamics, both theoretically and numerically (probability theory, Langevin equation, Fokker-Planck equation, ...). A previous experience in using computational methods for image analysis would be appreciated. The candidate should be willing to work closely with experimentalists, both physicists and biologists. The candidate will work in a multisciplinary and international environment, and will share his/her time between a biophysics lab and a physics institute.
Is this project the continuation of an existing project or an entirely new one? In the case of an existing project, please explain the links between the two projects
The theoretical modeling is a new project and a new collaboration. It is based on the recent experimental developments in LAI, including advanced image analysis (articles in preparation)
2 to 5 references related to the project
- Wolf, N. K., Kissiov, D. U. & Raulet, D. H. Roles of natural killer cells in immunity to cancer, andapplications to immunotherapy. Nat Rev Immunol (2022) doi:10.1038/s41577-022-00732-1.
- Alieva, M., Wezenaar, A. K. L., Wehrens, E. J. & Rios, A. C. Bridging live-cell imaging and next-generation cancer treatment. Nat Rev Cancer (2023) doi:10.1038/s41568-023-00610-5.
- Hamilton, P. T., Anholt, B. R. & Nelson, B. H. Tumour immunotherapy: lessons from predator–preytheory. Nat Rev Immunol 22, 765–775 (2022).
Two main publications from each PI over the last 5 years
- Limozin L, Bridge M, Bongrand P, Dushek O, van der Merwe PA, Robert P . (2019) TCR-pMHCkinetics under force in a cell-free system show no intrinsic catch bond, but a minimal encounterduration before binding. PNAS 116: 16943-16948 .
- Gonzalez C, Chames P, Kerfelec B, Baty D, Robert P, Limozin L (2019) Nanobody-CD16 catch bondreveals NK cell mechanosensitivity. BIOPHYSICAL JOURNAL 116: 1516-1526
- T. Guérin, N. Levernier, O. Bénichou, R. Voituriez,(2016) Mean first-passage times of non-markovian random walkers in confinement, Nature534, 356-359
- N. Levernier, J. Textor, O. Bénichou, R. Voituriez, (2020) Inverse square Levy walks are notgenericoptimal strategies in dimensions d >= 2 , Phys. Rev. Lett. 124, 080601
Project's illustrating image