PHD2024-03
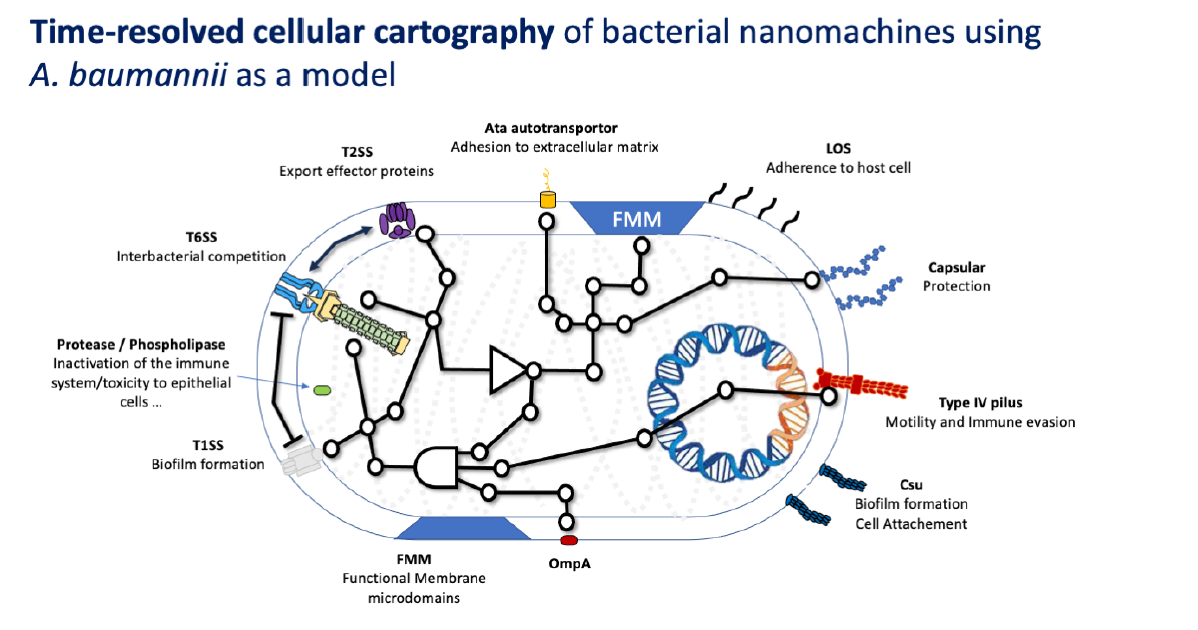
Cellular interactome of Acinetobacter baumannii virulence nanomachinosome: a multidisciplinary approach to decipher the paths, the tracks and their dynamics.
Host laboratory and collaborators
Eric Durand, LCB eric.durand@inserm.fr
Christine Brun christine-g.brun@inserm.fr, Andreas Zanzoni andreas.zanzoni@univ-amu.fr, TAGC, Inserm
Alain Schmitt, IGS schmitt@igs.cnrs-mrs.fr
Abstract
Bacterial cell envelope is densely packed with macromolecular apparatus, which are gigantic and energy consuming nanomachines some serving the pathogenesis while others the daily lifestyle of the bacteria. The PhD project will offer a paradigm shift by deciphering the molecular crosstalk that could orchestrate the building, positioning, and functioning of nanomachines. The project will explore the new concept of nanomachinosome: the ensemble of nanomachines, their relationship and dynamics in a changing environment. This project hypothesizes that the Acinetobacter baumannii virulence nanomachinosome is well organized within the cell envelope and relative to housekeeping cellular processes. Particularly, this equilibrium can be rearranged following environmental changes, leading to a second novel concept of “epinanomachinosome”. The project will unearth the basic principles governing nanomachinosome architecture and dynamic and determine how it plays a major role in the virulence of the pathogen and its capability to cause disease in human. Understanding the epi-nanomachinosome at molecular resolution is key to find new therapeutic avenues.
Keywords
Virulence nanomachines, bacterial pathogens, cellular microbiology, live-cell imaging, molecular microbiology, artificial intelligence, inference, modelling
Objectives
This PhD project seeks to decipher the dynamic network of interactions connecting the virulence nanomachines with housekeeping processes in the human pathogen A. baumannii with the aim to identify key nodes that are potential therapeutic targets. How virulence factors (VF) are integrated into the physiology of bacterial cells? Do VF hijack or shape house-keeping processes for their assembly/functioning? What is the adaptation of the nanomachine network in a changing environment?
Proposed approach (experimental / theoretical / computational)
1. Molecular to cellular cartography of A. baumannii virulence nanomachines (EXP.). We will record the presence, location and dynamic of VF using time-lapse fluorescence microscopy and quantitative data analysis.
2. Deconvolution through multilayer resolution (EXP.). We will quantify the (co)presence of the different VF using transcriptomic and quantitative proteomic in changing environments.
3. Genome wide in silico interactome of the T6SS nanomachine (COMP.). We will monitor structural connection between VF and house-keeping machines using artificial intelligence (AlphaFold) to perform in silico pull-down.
4. Experimental validation of the PPIN (EXP.). We will validate the protein-protein interaction network (PPIN) through the use of a nested combination of approaches (pull-down, fluo-microscopy, functional link using specific mutants).
5. Inference approach: refining the interaction domains (COMP.). We will use the mimicINT workflow to identify the interaction interfaces between the nanomachines proteins. We will use the biased Random-walk method to integrate quantitative proteomic data to better understand how the VF influences house-keeping machines in changing conditions.
Interdisciplinarity
This pioneering project combines for the first time, the biology of bacterial nanomachines (LCB) to AI-based prediction of the interaction network (IGS) with the computational approaches (TAGC). The project will integrate multi-method approaches from fundamental microbiology, to genetics and structural microbiology, macro- to micro- to nano-scale observations of bacterial nanomachine intimacy, in combination with AI-based prediction, inference and modelling. We will build a Nano-ATLAS describing the virulence nanomachinosome in the pathogen A. baumannii, their integration in the biology of the cell and the evolution of this network and its adaptative nature. The project will seek to discover a metanetwork that coordinates the virulence of A. baumannii – the epi-nanomachinosome – which is directly related to its incredible success as a human pathogen worldwide. In collaboration with Élisabeth Remy, we will integrate mathematical models to connect all the experimental data and to predict key nodes that control the nanomachinosome. This work will undoubtedly pave the way to develop new therapeutics targeting cornerstones of the Nano-ATLAS.
Expected profile
We seek for a candidate motivated by systems biology that includes computational modelling, AI-based approaches, data analyses as well as protein production and interaction studies. Previous experience in either bioinformatics and/or in biochemistry is expected. An interest in fundamental and molecular microbiology of bacterial pathogens will be a plus.
Is this project the continuation of an existing project or an entirely new one? In the case of an existing project, please explain the links between the two projects
The project is in continuation of a 4-years PhD project in the E. Durand team (grant DGA-AMU, end December 2023). The former PhD student (Ms Ona KANDOLO) initiated all the study on the Acinetobacter baumannii T6SS nanomachine. She set up the genetic approaches, the biochemistry and purification of difficult membrane protein complexes and their observation by cryo-EM, and the live fluorescence observation of nanomachine location and dynamics. In addition, a Master 2 student (AMU) developed the complementary in silico and in vivo pull down in collaboration with Dr Alain Schmitt (co-Supervisor 2).
2 to 5 references related to the project
- Allsopp, L. P., Bernal, P., Nolan, L. M., and Filloux, A. (2020). Causalities of War: The Connection Between Type VI Secretion System and Microbiota. Cell Microbiol. 22 (3), e13153. doi: 10.1111/cmi.13153.
- Harding CM, Hennon SW, Feldman MF. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat Rev Microbiol. 2018 Feb;16(2):91-102. doi: 10.1038/nrmicro.2017.148.
- Weber BS, Harding CM, Feldman MF. Pathogenic Acinetobacter: from the Cell Surface to Infinity and Beyond. J Bacteriol. 2015 Dec 28;198(6):880-7. doi: 10.1128/JB.00906-15.
- Gao M, Nakajima An D, Skolnick J. Deep learning-driven insights into super protein complexes for outer membrane protein biogenesis in bacteria. Elife. 2022 Dec 28;11:e82885. doi: 10.7554/eLife.82885. PMID: 36576775.
- Durand E, Nguyen VS, Zoued A, Logger L, Péhau-Arnaudet G, Aschtgen MS, Spinelli S, Desmyter A, Bardiaux B, Dujeancourt A, Roussel A, Cambillau C, Cascales E, Fronzes R. Biogenesis and structure of a type VI secretion membrane core complex. Nature. 2015 Jul 30;523(7562):555-60. doi: 10.1038/nature14667. Epub 2015 Jul 22. PMID: 26200339.
Two main publications from each PI over the last 5 years
Dr Eric Durand
- Kandolo O, Cherrak Y, Filella-Merce I, Le Guenno H, Kosta A, Espinosa L, Santucci P, Verthuy C, Lebrun R, Nilges M, PellarinR, Durand E. Acinetobacter type VI secretion system comprises a non-canonical membrane complex. PLoS Pathogens, 2023 Sep 28;19(9):e1011687. doi: 10.1371/journal.ppat.1011687. PMID: 37769028. hal-04233058.
- Cherrak Y, Rapisarda C, Pellarin R, Bouvier G, Bardiaux B, Allain F, Malosse C, Rey M, Chamot-Rooke J, Cascales E, Fronzes R and Durand E* (corresponding author*). Biogenesis and structure of a type VI secretion baseplate. Nature Microbiol. 2018 Dec;3(12):1404-1416. doi: 10.1038/s41564-018-0260-1. hal-02342924v1.
Drs Christine Brun, Andreas Zanzoni
- Kim D.K., Weller B., Lin C.W., Sheykhkarimli D., Knapp J.J., Dugied G., Zanzoni A., Pons C., Tofaute M.J., Kishore N., Sauer M., Rayhan A., Young V., Marín-de la Rosa N., Poirson J., Pogoutse O., Spirohn K., Strobel A., Laval F., Schwehn P., Li R., Rothballer S., Altmann M., Cassonnet P., Cote A.G., Vergara E.L., Hazelwood I., Liu B.B., Nguyen M., Pandiarajan R., Dohai B., Rodriguez Coloma P.A., Willems L., Twizere J.C., Taipale M., Jacob Y., Hao T., Krappmann D., Hill D.E., Brun C., Heinig M., Falter C., Aloy P., Demeret C., Vidal M., Calderwood M.A., Roth F.P. and Falter-Braun P. (2023) A proteome-scale map of the SARS-CoV-2 human contactome. Nature Biotech, 41:140-149, doi: 10.1038/s41587-022-01475-z.
- Veronika Young, Bushra Dohai, Thomas C. A. Hitch, Patrick Hyden, Benjamin Weller, Niels S. van Heusden, Deeya Saha, Jaime Fernandez Macgregor*, Sibusiso B. Maseko, Chung-Wen Lin, Mégane Boujeant, Sébastien A. Choteau*, Franziska Ober, Patrick Schwehn, Simin Rothballer, Melina Altmann, Stefan Altmann, Alexandra Strobel, Michael Rothballer, Marie Tofaute, Matthias Heinig, Thomas Clavel, Jean- Claude Twizere, Renaud Vincentelli, Marianne Boes, Daniel Krappmann, Claudia Falter, Thomas Rattei, Christine Brun, Andreas Zanzoni, Pascal Falter-Braun (2023) A gut meta-interactome map reveals modulation of human immunity by microbiome effectors. bioRxiv 2023.09.25.559292; doi: 10.1101/2023.09.25.559292. In review at Nature Microbiol. (centuri PhD students*)
Dr Alain Schmitt
- Villalta A, Schmitt A, Estrozi LF, Quemin ERJ, Alempic JM, Lartigue A, Pražák V, Belmudes L, Vasishtan D, Colmant AMG, Honoré FA, Couté Y, Grünewald K, Abergel C. The giant mimivirus 1.2 Mb genome is elegantly organized into a 30-nm diameter helical protein shield. Elife. 2022 Jul 28;11:e77607. doi: 10.7554/eLife.77607. PMID: 35900198.
- Rigou S, Schmitt A, Alempic JM, Lartigue A, Vendloczki P, Abergel C, Claverie JM, Legendre M. Pithoviruses Are Invaded by Repeats That Contribute to Their Evolution and Divergence from Cedratviruses. Mol Biol Evol. 2023 Nov 3;40(11):msad244. doi: 10.1093/molbev/msad244. PMID: 37950899.
Project's presentation video and illustrating image