CENTURI
Internship Program
Internship Program
CENTURI Internship Program
The Turing Centre for Living Systems (CENTURI) offers 3 to 6-month internships in research laboratories to highly motivated engineering students with backgrounds in Physics, Mathematics and Computational Sciences who are interested in applying their skills towards problems in Biology. CENTURI internship program is the opportunity for students to apply their skills in maths, physics and computer science to biological questions. The internships are funded and coupled with the CENTURI Summer School (June 20- July 01, 2022).
If you are interested in a project and wish to apply, please send an e-mail including your CV and a cover letter to the contact mentioned in the project.
Duration: 4 to 6 months (between January and September 2021)
Description of the host team:
The internship will take place in the Mann team "Axonal plasticity in development and cancer" at IBDM. The team studies how neuronal plasticity and function regulate cancer progression in mouse models of pancreatic ductal adenocarcinoma (PDAC). We have discovered that axons from different components of the peripheral nervous system innervate precancerous lesions and invasive cancers. We are also exploring how peripheral nerves may lead to accelerated or reduced tumor progression depending on their neurochemical properties.
Description of the environment:
The team is located at the IBDM (Institut of developmental Biology of Marseille), located on the campus of Luminy near the “Calanques”. The team has expertise in neuroscience, pancreatic cancer, 3D imaging on cleared tissue and image analysis. The team benefits from the Turing Centre for Living Systems (CENTURI), an interdisciplinary project that federates researchers in biology, physics, mathematics, computer science and engineering.
Description of the project:
Given the multi-faceted role of the different components of the peripheral nervous system in modulating PDAC progression, neuronal fibers represent an attractive target to curb tumor development and progression. Indeed, therapies tarteging neurotransmitters have been validated in mouse models, and some of these agents are currently under clinical evaluation. We are currently developing new approaches to target axonal remodeling and neuronal secretion. We are using 3D light sheet fluorescence microscopy to visualize in whole pancreas the architecture of neural networks, their interactions with the tumoral microenvironment and tumor progression. However, manual analysis of 3D images is a tedious and time-consuming task and quality of the analysed data can be low. The main goal of the project is to develop and use artificial intelligence techniques to automatically segment volumetric images of pancreatic tumors and quantifiy cancer growth and effects of treatments.
Interdisciplinarity and disciplines involves:
- Deep learning based auto-segmentation
- biology
- pancreatic tumor segmentation
- therapy of pancreatic cancer
- 3D cleared image analysis.
Expected profile:
Skills in artificial intelligence with knowledge in software development, data science, data engineering and programming are wished. Depending of the expertise of the candidate, the project could be adapted by using images analysis methods already present in the team.
Contact:
Sophie Chauvet: sophie.chauvet@univ-amu.fr
Duration: 6 months
Description of the host team:
The internship will take place in the team "Neural Bases of Sensorimotor Learning" . The team's projects aim to improve our understanding of the biological algorithms that contribute to behavioral adaptability and variability, with a focus on how individual and species-specific sensitivity to costs (cost of movements and of time) affect both decision-making and motor control.
Description of the environment:
The internship will take place at INMED, an internationally renowned Neuroscience research center, bringing together 9 research teams and around 150 people (http://www.inmed.fr). The Institute is located in the Luminy Campus at the entrance of national park of the Calanquens. INMED is a unique place of scientific synergy, bringing together research teams with complementary expertise to study the function and pathology of the brain
Description of the internship project:
Imagine yourself stepping on a long escalator. If you are in a hurry (i.e., sensitive to time), you will spend energy to quickly climb the stairs and reach the top of that escalator as early as possible. Alternatively, if you are exhausted (i.e., sensitive to effort), you may decide to stand still while being carried upward, even if that means arriving later at your final destination. Thus, many of our reward/goal-oriented decisions and actions reflect a tradeoff between effort and a cost associated with the passage of time. Still, how humans and animals trade effort against time in the natural context of foraging-like behavior when both effort and time are experimentally manipulated, has not been investigated comprehensively. The goal of the project is to develop a behavioral apparatus for rodents to study their foraging strategy (exploit or explore) while manipulating effort and cost of time.
Interdisciplinarity and disciplines involves:
Mechatronic (electronic, automation, python-based programing , signal processing)
Expected profile:
Capability to work at the interface software/hardware to acquire data from sensors (infrared-beam, camera) and control actuators (solenoid valves, motors). For instance experience or motivation to work with Arduino, raspberryPI, python will be a plus. Strong motivation to learn
Contact:
David Robbe: david.robbe@inserm.fr
Duration: 6 months
Description of the host team:
We are a team of neuroscientists interested in genes and brain circuits altered in neurodevelopmental disorders. We use mouse and rat models of disorders. Part of the team studies the pathophysiology of epilepsy, while another part focuses on autism and disorders of social interaction and memory.
Description of the environment:
INMED (http://www.inmed.fr/) is a prestigious neuroscience research center located in the spectacular scenery of the calanques national park. It hosts 9 teams focused on understanding brain function and plasticity in health and neurological diseases. In particular, INMED researchers have pioneered the technique of calcium imaging in vivo, thereby forerunning our understanding of neural network dynamics in living animals.
Description of the internship project:
We have recently acquired 2 Inscopix nVoke miniature microscopes. These are high quality miniaturized head-mounted microscopes that, combined with brain-implanted lenses, provide the possibility to record the real time activity of 500-1000 neurons in deep brain regions of freely moving mice for months. We are currently using these tools to study cell ensemble dynamics in social memory circuits (the ventral CA1 of the hippocampus) in mice, both in normal conditions and in a genetic model of autism. More specifically, we record simultaneously neural activity and mouse social interactions and then correlate the two recordings to infer causal relationships. It was recently shown using Inscopix microscopes that there are ‘social cells’ in the mouse hippocampus, active only when the animal interacts with a specific partner. Each conspecific mouse has its own ‘social cell’ signature in the brain of the analyzed animal, so that we can ultimately predict which partner the animal is interacting with simply by observing neural activity in its ventral CA1.
The first aim of the proposed project aims at studying the activity of social cells in greater detail (synchronicity, ensemble configuration changes during familiarization with a conspecific, contribution of different neuron types to the social signature) during a social interaction test (in the 3 chamber assay, where the test mouse can choose to interact between a familiar and a novel conspecific). Our second aim is to study how ventral CA1 social cell networks are altered in our genetic mouse model of autism, the Neurod2 KO mouse. This type of project typically requires a large part of bioinformatics analyses.
Interdisciplinarity and disciplines involves:
Bioinformatics, computation, Matlab and Python coding, Neural networks
Expected profile:
We seek for a talented and highly motivated student with skills in bioinformatics and interest in brain imaging in freely moving mice and neuropsychiatric disorders. Good knowledge of Matlab will be an important.
Contact:
Antoine de Chevigny: antoine.de-chevigny@inserm.fr
Duration: 6 months
Description of the host team:
The Rancz laboratory is moving from The Francis Crick Institute in London to Inmed in Marseille in December 2021. We are studying information processing in the mouse brain and build cutting edge research tools. The intern will be supervised by the PI, Ede Rancz, and will work closely with a neuro-engineering company, NeuroGears, the developers of Bonsai-Rx.
Description of the environment:
Inmed is a leading neuroscience institute located at the Luminy campus in Marseille. Several laboratories study animal navigation and behaviour using cutting edge research tools, creating an inspiring environment for biologists and engineers alike. The intern will have access to a dedicated 3D printer (Form 3), and basic electronics and mechanical workshops on site, as well as fabrication workshops on campus.
Description of the internship project:
Recording brain activity in behaving animals is essential to understand cognition. The more we can control the physical environment during these recordings, the more we can learn about internal brain processes. The project builds on a currently available, novel research device developed by us to control the visual and vestibular environment for navigating animals. This device allows head-fixed mice to locomote on a rotational platform driven via a Teensy micro-controller. Additional modules on the platform include machine vision cameras and biological recording devices. The main goal of the project is to integrate the current platform with newly developed recording and stimulating devices and implement system control under the Bonsai-Rx.org environment. Depending on the skillset of the intern, the emphasis will be on mechatronic development of new modules, system control or implementation of visual virtual reality.
Interdisciplinarity and disciplines involves:
Mechatronics, Control system engineering, Computer Science
Expected profile:
We are looking for a curious, driven, and highly able engineering student. Experience with 3D CAD software (Solidworks, Autodesk Inventor or similar) and rapid prototyping, as well as good command of English, are essential. Working knowledge of control theory (PID controllers, motion control), VR engines (especially Vulkan) or single-board computers (e.g. Raspbery PI) is desirable.
Contact:
ede.rancz@inserm.fr
Duration: 4-6 months
Description of the host team:
This project is developed in collaboration between the LAI and the LMA. The LAI develop and apply advanced force spectroscopy techniques to unravel the physics behind biological systems, from single molecules to membranes and living cells. We are notably using acoustic force spectroscopy, an emerging technology that needs to be developed to match our own applications. The "Waves and Imaging" team at the LMA is specialized in the development on non-invasive ultrasound techniques for the characterization and diagnostic imaging of biological tissues (bone, tumor, blood).
Description of the environment:
We are developping experimental approaches and applying physical concepts to achieve a quantitative understanding of cellular mechanics from a fundamental point of view but with an eye on applying our approches to the biomedical field.
Our interdisciplinary and international groups integrate members coming from physics, engineering, and biology backgrounds.
Description of the internship project:
Acoustic force spectroscopy (AFS) is an emerging technique that use acoustic forces in a microfluidic channel containing suspended cells to probe several cells in parallel (contrary to other methods that provide limited statistics on single cells) [1]. The commercial setup is not fully matching our needs (lack of calibration, manipulation of cell clusters). The aim of the project is to better characterize the current setup (precise characterization of the acoustic field within the microfluidic channel using hydrophones) and to develop a new acoustic chamber. Alternative configurations (like single acoustic tweezers [2]) could be considered for the development of this new setup that should enable to manipulate bigger elements, such as cell clusters. This would enable to unveil their rheology (Young modulus, adhesion, yield-stress) and predict their behavior in complex hydrodynamic environments.
References:
Sitters et al. Nature Methods, 2015.
Lim et al. Microsystems & Nanoengineering, 2020.
Interdisciplinarity and disciplines involves:
Physics (acoustic, mechanics), Engineering
Expected profile:
We are looking for a student with background in physics or engineering and an interest in technological development. Experience in acoustic will be appreciated. The internship could be potentially followed by a PhD thesis.
Contact:
claire.valotteau@inserm.fr ; franceschini@lma.cnrs-mrs.fr
Duration: 3 to 6 months
Description of the project:
TheDuring embryonic development, cells acquire different fates that will give rise to different organs and tissues. A key example is gastrulation, which results in formation of the germ layers and the main body axes. This process involves collective movements of cells upon early cell differentiation. A puzzling question is how these early differentiation patterns form. It relates to the fundamental problem of how cells in an embryo determine their position relative to each other, and to the whole embryo. To explore these questions in the context of early mammalian embryos, we use aggregates of mouse-embryonic stems cells, so-called gastruloids. These in vitro models of early embryos show a fascinating capacity to self-organize and establish a primary axis and germ-layer-like patterns from an initially equivalent mass of cells. One hypothesis of how these patterns arise is that cells sense signaling molecules (“morphogens”) that are released by other cells or penetrate from the culture medium into the intercellula space. However, we currently lack tools to detect the low concentrations of such molecules directly in living embryos.
The objective of this interdisciplinary project is to employ quantitative fluorescence microscopy techniques such as fluorescence correlation spectroscopy (FCS) to detect diffusion of molecules inside embryonic organoids such as gastruloids. Applications of these techniques have so far been limited to isolated cells or small multicellular specimen, since aberrations and scattering distort the excitation volume in larger specimen and reduce the signal detected from
deeper sections. This projects aims to circumvent these limitations and to establish FCS as a tool applicable in organoids. In particular, we will implement FCS on a single-objective lightsheet microscope (soSPIM) and evaluate several correction schemes for sample-induced aberrations and scattering. We will compare the performance of soSPIM-FCS to confocal and 2-photon microscopes, and benchmark it directly in gastruloids. Once established, we will apply the developed tools to quantify the diffusion dynamics and concentration of fluorescently labeled morphogens inside gastruloids and correlate the detected local concentrations with cell differentiation and patterning.
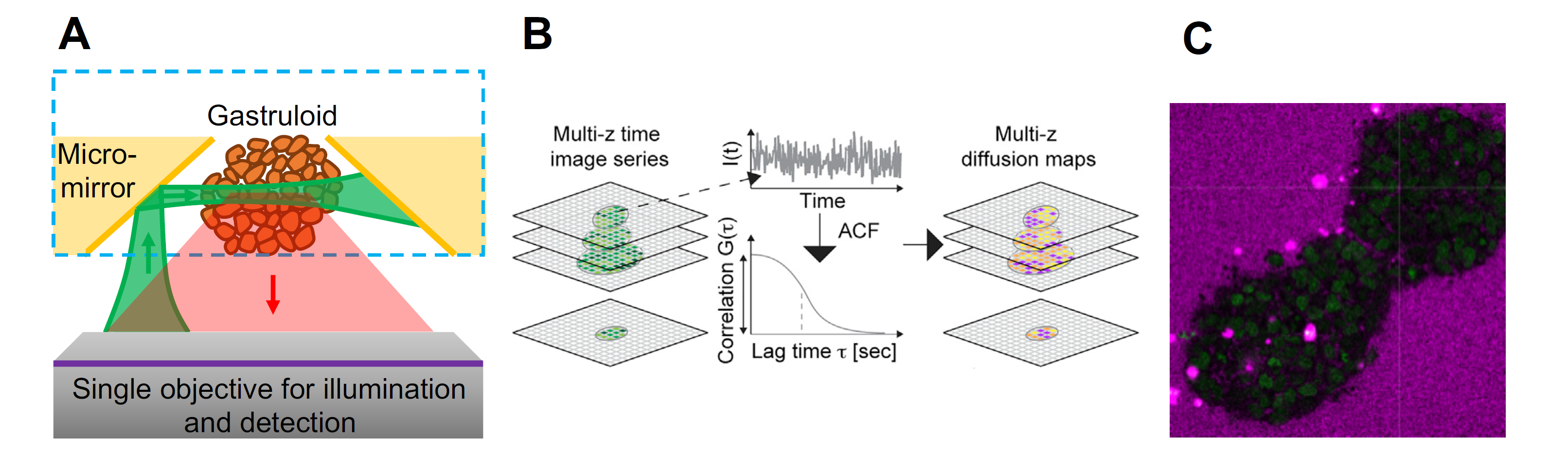
Figure 1. (A) Scheme of soSPIM setup. (B) soSPIM-FCS : 3D diffusion maps are generated by pixel-wise fluctuation analysis of soSPIM imaging stacks (from Singh et al., BJ 2017) (C) Image of gastruloid expressing nuclear marker H2BGFP (green) and extracellular dye sulforhodamine (SR, magenta). The cross marks the position of confocal FCS of SR. The project requires a solid quantitative background, basic knowledge in optics and strong interests in fluorescence microscopy and living systems. The project will be carried out in a biophysics lab that combines experimental and theoretical approaches to investigate different aspects of morphogenesis in several model organisms. This project may lead to a PhD.
Interdisciplinarity and disciplines involves:
complex systems, advanced microscopy, image analysis, developmental biology, physical models
The project requires a solid quantitative background, basic knowledge in optics and strong interests in fluorescence microscopy and living systems. The project will be carried out in a biophysics lab that combines experimental and theoretical approaches to investigate different aspects of morphogenesis in several model organisms. This project may lead to a PhD.
Contact:
Pierre-François Lenne and Valentin Dunsing: Pierre-francois.lenne@univ-amu.fr; valentin.dunsing@univ-amu.fr
